* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 2: Cellular signalling and cell division
Biochemical switches in the cell cycle wikipedia , lookup
Tissue engineering wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell encapsulation wikipedia , lookup
Endomembrane system wikipedia , lookup
Cell culture wikipedia , lookup
Cell growth wikipedia , lookup
Cytokinesis wikipedia , lookup
Programmed cell death wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cellular differentiation wikipedia , lookup
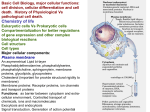
1. Basic Cell Biology, major cellular functions: cell division, cellular differentiation and cell death. History of Physiological Vs pathological cell death. Chemistry of life Eukaryotic cells Vs Prokaryotic cells Compartmentalization for better regulations of gene expression and other complex biological reactions Cell structure Cell types Major cellular components: 2. Cellular signaling and cell division 3. Cellular Differentiation and development, Cell death: history of progress in cell death research, necrosis vs. apoptosis. Basic Cell Biology, major cellular functions: cell division, cellular differentiation and cell death. History of Physiological Vs pathological cell death. Chemistry of life Eukaryotic cells Vs Prokaryotic cells Compartmentalization for better regulations of gene expression and other complex biological reactions Cell structure Cell types Major cellular components: Plasma membrane An asymmetrical Lipid bi-layer Phasphatidylethenolamine, phasphatidylserine, phasphatidylcholine, sphingomylein, membrane proteins, glycolipids, glycoproteins Cholesterol (important for provide structural rigidity to membrane) Membrane proteins: Receptor proteins, ion channels and transport proteins Functions: as barrier between cytoplasm and extra-cellular environment Controlled transport of chemicals, ions and macromolecules Exocytosis and endocytosis Signal transduction Generation of action potential Endoplasmic reticulum: Lipid bilayer forming a continuous sheet enclosing a single space called ER lumen or cisternal space Rough and smooth ER Functions: Protein synthesis Post-translational modification (glycosylation Glycolipd synthesis Vesicular Protein transport and secretion Detoxification (Smooth ER contain cytochrome P450), water insoluble toxic compounds are converted into excretable non-toxic soluble compounds Ca2++ sequestering: Examples- muscle cells (ER is called sarcoplamic reticulum), nerve cells. Golgi bodies: Membrane bound flattened sacs stacked over each other. Functionally distinct parts (cis and trans parts) Contains protein modification enzymes e.g. glycosyl transferase, nucleoside diphosphatase and acid phosphatase Functions: Posttranslational modifications (glycosylation, dephosphorylation, phosphorylation) Involved in sorting and packaging macromolecule for secretion or for delivery to other organelles. Proteins destined for delivery to lysosomes are labelled with mannose-6phospate in Golgi bodies. Defect in this process results in lysosomes without hydrolytic enzymes and secretion of these enzyme in I-cell disease or inclusion cell disease). Lysosomes: Membrane-bound vesicles containing hydrolytic enzymes Function: Involved in intracellular digestion Peroxisomes: Membrane-bound vesicles containing oxidative enzymes such as catalase and urate oxidase, and long chain fatty acid oxidation during which there is production of hydrogen peroxide. Functions: Vestige of an ancient organelle that carried out oxidative reactions May play role in anti-oxidative defence Mitochondria: Double membrane-bound structures, power plants of eukaryotic cells Site for citric acid cycle and oxidative phosphorylation and fatty acid metabolism in energy generating biochemical pathway Outer mitochondrial membrane Inter-membrane space Inner mitochondrial membrane (contains components of electron transport chain, ATP synthase, and transport proteins) Matrix space (contains enzymes and cofactors for citric acid cycle, fatty acid metabolism, mitochondrial genome, and transcription and translation machinery) Inner mitochondrial potential is generated and maintained by proton export out side ATP is synthesis is driven by proton flow towards inside Protein import into mitochondria Nucleus: Nuclear envelope Chromatin Replication, transcription Protein import and RNP export across nuclear envelope Lecture 2: Cellular signalling and cell division Cellular signalling: Evolution of social behaviour in cells Cell to cell communication and responses are essential for the organism as whole. Different types of cell signalling: synaptic Endocrine Paracrine Autocrine Cell to cell signalling by direct contacts: a) via receptors b) via gap junctions and plasmadesmata Extra-cellular signals: Hormones Cytokines Growth factors Signalling Mechanisms: 1. Receptor enzyme mediated 2. G-protein linked Receptor mediated 3. Ion-channel-linked Receptor mediated 4. Intra-cellular Receptor mediated Receptor enzyme mediated: Receptor tyrosine kinases Examples- receptors for most of growth factors e.g. FGF, PDGF, Insulin, IGF1, CSF Binding of ligand to RTK----activation of TK activity------autophosphorylation----binding of GTPase activating protein or PI3 kinase or phospholipase---Activation of PKC and/or Ras---activation of MAPKKK----activation of MAPKK---Activation of MAPK----c-Jun---activation of transcription Receptor serine/threonine kinases: Examples- TGF-family of receptors G-protein linked receptor mediated Signalling: Examples- adrenalin,Calcitonin, oxytosin, acatylcholine, dopamine, histamine, PTH, retinal, serotonin etc. Binding to receptor -----conformational change in G-protein complex-----GGTP formation-----activation of adnylate cyclase-----production of cAMP--activation of protein kinase A (camp dependent kinase)----- Ion channel-linked Receptors: Neurotransmitter receptors NMDA receptors, serotonin, acetylcholine receptors etc Binding----opening of ion channel------influx of Na, or K or Ca ions---downstream events Drugs: barbiturates, antidepressants used a blockers Intra-cellular Receptors Mostly steroid hormone receptors: Cortisol, estrogen, progesterone, thyroid hormone, retinoic acid, vitaminD Binding to receptor---- exposure of DNA-binding site, and NLS---activation of transcription NO signalling: Recently discovered as signalling molecule. Diffuses through plasma membrane rapidly and binds to Gunylate cylase. Binding to heme group of gunylate cyclase Activation production of cGMP activation of downstream enzymes Causes muscle relaxation, involved in release of neurotransmitters, at high level neurotoxic. Cell division: The fundamental principle of life Interphase: cells look normal and just grow in size as observed under microscope. But many molecular events take place in this phase and this phase further subdivided into following subdivisions: G1 phase: Starts after completion of mitosis and ends before beginning of DNA synthesis. Important decision to start cell cycle is made in the late G1 phase. S phase: This phase is marked by the start and completion of DNA replication. G2 phase: Cells prepare for cytoplasmic events for cell division and make sure that there are appropriate conditions before entering the M phase. Mitotic phase: Visible changes in the cells Check points in cell division cycle Cell cycle control: M-phase promoting factor – a complex of ser/thr kinase called cyclin-dependent protein kinase Cdk-2 or cdc2 and a mitotic cyclin Synthesis and destruction of cyclins control the activity of MPF. Re-replication block Growth factors trigger cascade of intracellular signals Growth factor binding---activation of TK-----activation of MAPKKK-------transcription of myc gene-----transcription of Cdk and cyclins Mitosis Prophase Metaphase Anaphase Telophase Cytokinasis Remember that DNA replication takes place in prophase and DNA is packed into a condensed form called chromosomes. Each chromosome has two chromatids, and during metaphase and telophase, each chromatid separates and daughter cells have the same number of chromosomes. Meosis: Prophase I---subdivided in to 5 stages Leptotene---Chromatids are formed, Chromosomes look like treads Zygotene----Homozygous chromosome come close forming pairs Pachetene---further condensation of chromosomes Diplotene---Completion of pairing of chromosomes (completely aligned chromosomes) Dikinasis---chromosomes separate, and chromatids are attached at the places of crossing over or chiasma. (chromosomes look like garlands) Metaphase I Anaphase I (separation of chromosomes and not the chromatids) Telophase I Meotic division II: same as usual mitosis Lecture-3/4 Cellular Differentiation and development, Cell death: history of progress in cell death research, necrosis vs apoptosis. Control of gene expression: All the cells in an organism contain the same genetic information. Cells differ from each other not because they contain different genes but because they express different genes. Different steps of control of gene expression: Transcriptional control RNA processing control RNA transport control Translation control MRNA degradation control Protein activity control Molecular regulators of gene expression at transcriptional level Transcription activator Special class of DNA binding proteins Transcription repressors Four common structural motifs: Helix turn helix motif: Two helices connected by a short extended chain of amino acids which makes the turn. Zn-finger motif: Consists of an -helix and a -sheet held together by Zn (coordination bond with four amino acid side chains) Lucine Zipper motif: Two -helices (one from each monomer) joined to gather to form a short coiled coil by hydrophobic interaction of hydrophobibic amino acid side chains (mainly lucine) of two helices. Helix loop helix motif: A short -helix connected by a loop to another long helix (loop provide flexibility of the two helices) These proteins generally have two domains: one which binds to specific DNA sequence and other which alters the activity of RNA polymerase (or transcription assembly) Example of prokaryotic genetic switch: lac-operon: first gene in this operon encodes -galactosidase, an enzyme required for digestion of lactose producing glucoase and galactose Glucose cAMP inactive CAP ------no transcription No glucose------low camp-------Active CAP----Gene is transcribed if lactose is available in medium (lac repressor is not bound) Lactose-----allolactose------binds to repressor protein------release of repressor from DNA -----transcription is allowed if CAP is bound Eukaryotic Transcription is complex: requires many transcription factors in addition to RNA polymerase II Control of gene expression during development: In amphibian development early signals for differentiation are due to the asymmetry of the zygote. Transplantation experiment showed the dorsal lip of the blastopore initiates gastrulation movement which organizes the body plan. For example Vg1 mRNA is synthesized in the oocyte and become localized in vegetal cortical region of egg. An injection of mRNA coding the active Vg1 protein in the ventral blastomere can induce an entire body axis. Morphogens: the biochemical species responsible for differentian The gradient of morphogen can organize complex pattern as cells respond differently to different concentrations of morphogen Mammalian Development: in protected uterine environment Nutrient provided by host through placenta The embryo proper is derived from inner cell mass The trophectoderm is the precursor for placenta Totipotent cells: cells of the early mammalian embryo (up to eight cell stage) are identical and unrestricted in their capabilities. Capable of developing in normal animal. Mammalian embryonic stem cells: Cells of the inner cell mass can be dispersed and grown in culture under appropriate condition. They divide indefinitely without any change in character. They are used in making transgenic animal as well gene-knockout transgenic mutants. Caenorhabditis elegans: Very simple multi-cellular organism Transparent body, every cell has been studied in terms of its origin during development and its function. Total 1090 somatic cells 131 cell die by apoptosis during development 14 genes have been identified, which affect the programmed cell death when mutated. They are called Ced-1, Ced-2, Ced-3, Ced-4------Ced-14. CED stands for cell death defective. Mammalian Homologues of Ced-3, Ced-4 and Ced-9 have been discovered. These three genes have been studied extensively and they play critically important role in apoptotic cell death. Ced-3= ICE or Caspase-1 Ced-4= APAF-1 Ced-9= Bcl-2 Cell death: Pathological or necrotic cell death Programmed cell death or Apoptosis Initial experiments by Dr Kerr Liver ischemia Morphology: shrinking cells, membrane intactness, chromatin condensation Apoptotic bodies Change in lipid composition of plasma membrane (flipping of negatively charged phosphoserine or negatively charged lipids from inner leaflet to outer leaflet). Increase in transglutaminase activity-leading to cross-linking of proteins and lipids Necrosis: pathological, accidental and traumatic: cell swelling, membrane breakage, release of lysosomal enzymes in the vicinity, causing inflammation, damaging the nebouring cells. Biochemical Feature: DNA fragmentation: In apoptosis –oligonucleosomal fragmentation, leading to DNA ladder In necrosis: no damage or random fragmentation leading to smear Apoptosis during development: metamorphosis, development of digits, massive death of neurons, eye development Apoptosis in adult tissue: Intestine, skin, thymocytes, uterus cells, infected, transformed or damaged cells Lecture5/6 Apoptotic cell death during developmental and during adult tissue homeostasis: A. Development: Genetic basis of cell death in C. elegens development, examples of PCD in mammalian neuronal development, and inter-digital cell death during development and metamorphosis. B. Tissue homeostasis: Cellular homeostasis in blood cells, immune system, epithelial cells, hormone dependent changes in tissue architecture such as those in breast and uterine tissues. C. Physiological cell death as safeguard against growth of cells carrying mutations (potentially capable of becoming cancerous) Example of programmed cell death during development: a) C.elegens development: ced-3 and ced-4 genes encode pro-apoptotic proteins. These proteins are required for cell death. Mutation in these genes led to the prevention of death of 131 cells of the c. elegeans and the worm developed unusually but survived. But if the ced-9 gene was defective, all the cells died and so did the embryo. The ced-9 encodes an anti-apoptotic protein. It is required in the other cells to prevent cell death (Ellis R E and Hoevitz (1986) Cell, 44: 817-819, Yuan, J and Horvitz, H. R. (1990) Dev. Biol. 138: 33-41). b) In vertebrates: Metamorphosis: Cells in the tadpole tail are triggered to die by thyroid hormone secreted by thyroid gland. Similarly the intestine of tadpole is totally degenerated by apoptosis induced by Thyroid hormone. Apoptosis can be induced by adding to the epithelial cultures from tadpole. TH responsive gene expression----matrix metalloproteases were expressed in response to TH hormone (Patterton et al. (1995) Devel. Biol. 167:252-262). Neuronal cell death during development (Johnson EM and Deckwerth TL (1993) Annual Rev. Neurosci. 16:31-46). Peptide inhibitors of caspase-1 arrested apoptosis of motor neurons in vivo (Milligan et al, 1995, Neuron, 15 : 385-393) Transgenic caspase-3 knockout mice, died post-natally (Kuida et al (1996) Nature, 384: 368-372. Large scale astrocyte death in developing cerebellum More than 50% oligodendrocytes die during developing rat optic nerves. Culture of oligodendrites require factors secreted by optic nerve cells. They could be rescued by PDGE or IGF-1(Barres et al 1992, Cell 70:31-46). Other culture models: Differentiation of PC-12 cells into neuronal cells by NGF. The differentiated cells depend on NGF for survival. Removal of NGF triggers apoptosis in these cells. Tissue homeostasis: Requirement of survival signals: Prostrate cells require testosterone to survive. Cells in adrenal cortex depend on ACTH secreted by pituitary gland. Interlukin-2 dependent T- lymphoblast cells require IL-2 for survival Endothelial cells require fibroblast growth factors (FGF) for survival Game of cell proliferation and cell death in Immune system: T lymphocytes: Responsible for cell mediated immunity Cytotoxic T cells or killer T cells Helper T cells: produce IL-2, and activate other lymphocytes B lymphocytes: Mount antibody response against antigens Lymphocyte maturation in Thymus Cells expressing no receptor and cells expressing high affinity to self antigens are induced to undergo apoptosis Cells expressing receptors for foreign antigen and low affinity for self antigens survive and are released in circulation- resting T cells When they encounter antigen ----they get actvated-----proliferate, synthesize lymphokines, produce perforin and grazyme B as well as non-functional Fas receptor and ligand This leads to removal of antigen After the antigen is gone---now unnecessary cells are induced to undergo apoptosis by 1. Deprivation of IL-2 survival signal or 2. by making functional fas receptor and ligand, and inducing cell death