* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Antibody structure : the early studies
Epigenetics in stem-cell differentiation wikipedia , lookup
Minimal genome wikipedia , lookup
Copy-number variation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
DNA vaccination wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
X-inactivation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genome evolution wikipedia , lookup
Point mutation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
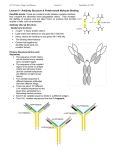
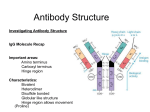
Antibody structure : the early studies Albumin Globulins Untreated antiserum Absobanc A Ab rbance bsor c Antiserum upon p incubation with the Ag Electrophoretic migration Antibody structure : the early studies (cont.) Fab Fab papain Fc F(ab')2 pepsin pFc pFc' Light chain reduction Heavy chain Antibody structure by electron microscopy Angle between arms is 0 degrees Angle between arms is 90 degrees Angle between arms is 60 degrees Antibody arms are joined by a flexible hinge How Antibodyy Binds to Antigen g The top part of this figure shows how different shaped antigens can fit into the binding site of antibodies: left, pocket; center, groove; right, extended surface. The panels below show space-filling or computer-generated t t d models d l iindicating di ti where h contact t tb between t th the peptide antigen and antibody occurs. How an Antibody Works When an Ab finds its Ag on an invader, it will bind there and act as a “trash tag”, marking it for destruction by “killer” cells, ll macrophages h or complement l Antibody binds to target antigen Receptor for constant region of antibody on NK cell recognizes a bound antibody After binding, the NK cell is signaled to kill the target cell The target cell dies by apoptosis and/or membrane damage Antibody structure by modern techniques Antigenbinding sites Antigenbindingg sites VH VH CH1 CH1 VL h hinge disulfide bonds carbohydrate hinge g CL CH2 CH3 VL CL carbohydrate CH2 CH3 disulfide bonds Diagrammatic IgG structure Light Light chains chains Antigen binding site hinge Papain Pepsin carbohydrate carbohydrate Heavy chains Immunoglobulin G (IgG) • L chains have 2 domains each (VL and CL). • H chains of IgG (IgA and IgD also) have 4 domains (VH, CH1, CH2, CH3). • The Hinge region are cys- and pro-rich; it is flexible, open accessible and located between CH1 and CH2 open, domain. • There are carbohydrate moieties attached to the CH2 domain in the two H chains. • Pairing: VL/VH, CL/CH1, CH2/CH2 and CH3/CH3. • H chains of IgM and IgE have 5 domains (VH, CH1, CH2, CH3 and CH4). 4) There is no hinge region in these two isotypes. Antibody architecture IgD IgM IgG IgE IgA κ or λ μ γ δ α ε Heavy chain constant domains determine isotype J chain J chain IgM and IgA can form polymers Light chain (approx. 25 kDa) Two types of L chains: Kappa (κ) and Lamda (λ) encoded by genes located in different chromosomes. Ratio of κ to λ chains can vary (mouse: 20/1 κ/λ; human: 2/1 κ/λ, cattle: 1:20 κ/λ). The two L chains of any one Ig molecule contain either κ or λ, λ i.e. i e ne never er one κ with ith the other λ. λ Heavy chain (approx. (approx 50 kDa) Five classes or isotypes of antibodies defined by the H chains: Ig class (isotype) H chain IgM μ IgG γ IgA α IgD δ IgE ε The five H chains differ in their primary structures with approx. 60% homology. Four subclasses (subtypes) of IgG: Human: IgG1 (γ1), IgG2 (γ2), IgG3 g (γ (γ3)) and IgG4 g (γ (γ4). ) Mouse: IgG1 (γ1), IgG2a (γ2a), IgG2b (γ2b) and IgG3 (γ3). Two IgA subclasses (IgA1 (α1) and IgA2 (α2)) (α2)). • Extensive E t i h homology l ((approx. 90%) among th the subclasses b l off H chains that belong to the same isotype. • Both H chains are identical in any one Ig molecule. Reading: Chapters 3 and 4. Antibody fine structure Light chain C domain Light chain V domain beta strands beta strands disulfide bond A Arrangement t off bbeta t strands t d Antibody fine structure (cont.) (cont ) CDR Heavy-chain V region Light-chain V region HV3 HV2 HV1 Variability HV3 HV1 HV2 Variabilityy FR1 FR1 FR2 FR2 FR3 FR3 Residue # HV : hypervariable region (= CDR : complementarity determining g ) region) FR : framework region Residue # • Three Hypervariable regions, HV1, HV2 and HV3, present in each of the H and L chains. • They are franked by less variable regions called Framework regions, FR1, FR2, FR3 and FR4. • The combination/folding in HV regions bet between een H and L chains constitutes the antigen binding sites that are complementary in structure to the Ag epitope, termed complementarity-determining regions (CDRs), CDR1, CDR2 and CDR3. The a.a. a a in the CDRs provide multi-point contacts and different types of interaction with those in the Ag epitope: H-bonds, H-bonds electrostatic forces, forces hydophobic forces and van der Waals forces. Together they provide stable noncovalent Ab/Ag association. Antibody fine structure (cont.) Variability N C residue # N C Ag-binding site N C The nature of antigen-antibody interactions Antigen Antibody Gln Ser Lys Hydrogen y g bond Ionic bond F~1/d 7 Glu Val Val Leu Asp Hydrophobic bond & Leu Van der Waals interactions Lys Ionic bond F~1/d These are allll REVERSIBLE forces. Th f They only act on SHORT DISTANCE. Th can yield They i ld either ith attracion tt i or repulsion. li 2 Like a key in a lock poor fit good fit Ab Ab Ag Ag high attraction - low repulsion high repulsion - low attraction Ab Ag Ab Ag affinity =Σ attractive and repulsive forces Affinity Affi it M Maturation: t ti The Ag-specific abs produced in secondary response exhibit greater affinity than those seen in the primary response. Antibody-Antigen interaction : a dynamic equilibrium association (k1) Ab + Ag AgAb dissociation (k-1) K = k1/ k-1 K = [AbAg] /[Ab] [Ag] (association constant) If [[AbAg] g] = [Ab], [ ], that is,, if half of all the Ab is occupied, K = 1/[Ag] ( dimension of K: liter per mole) If K iis hi high, h lilittle l A Ag iis needed d d to occupy hhalf lf off the h Ab Ab. p on the reaction conditions! K is dependent Affinity determined by equilibrium dialysis % labeled antigen distribution semipermeable membrane A B A without antibody B % with antibody antigen g bound by antibody Initial state antibody labeled antigen Equilibrium Affinity determined by equilibrium dialysis (cont.) Equilibrium dialysis of diffusible antigen Add Ab to low Ag concentration No antibody concIN = concOUT Add Ab to high Ag concentration concIN = concOUT + concBOUND Scatchard analysis Single antibody Mixture of antibodies Mixture of Abs Mixture of Abs r conc of free Ag slope = - KA Moles Ag bound per mole Ab (= r) Affinity and Avidity Fab IgG IgG IgM Eff ti Ab valence Effective l 1 1 2 Antigen valence 1 1 n n Association const. 104 104 107 1011 Cooperative advantage - - 103 107 up to 10 Definition of binding intrinsic affinity avidity or functional affinity Applications liquid-phase techniques solid-phase techniques Antibody Variability There are several reasons why there are an enormous number of different antibodies: • different combinations of heavy and light chains which are encoded by different genes • recombination • others The Number Dilemma • You have about a trillion different antibodies able to react with millions of different types of Ag • b butt you only l have h about b t 30,000-40,000 30 000 40 000 genes which hi h code d for f all ll the proteins you need in your entire body, most of which are not Ab • so there cannot be one gene for one antibody to code for these – we wouldn’t have enough antibodies! S hhow can your bbody So d produce d Ab to so many antigens, i even those h it’s never seen? Antibody Genes Genes for antibodies aren’t like most other genes - they come in pieces ((“gene gene-lets lets”): ): • • • • variable i bl segments (V) ( ) – many different diff versions i diversity segments (D) – several different versions joining segments (J) – a few different versions constant segments (C) – a few different versions that are nearly l identical id ti l Other sources of variability • when V, D, and J pieces are joined, they may not always be joined perfectly – if some base-pairs base pairs are lost or added, added the Ab will end up with a different amino acid sequence • variable region genes mutate at a higher rate than other genes in your body Lecture 7 to 9 (cont.II) Generation of Antibody Diversity Overview: Gene rearrangement in the generation of antibody diversity. Junctional and insertional diversity. Somatic hypermutation. Allelic exclusion Genes encoding g human κ L chain ((chromosome 2)) λ L chain (chromosome 22) H chain (chromosome 14) The genes encoding the H and L chains are arranged in exons separated by the intervening introns. L chains have gene segments of V (variable), (variable) J (joining) and C (constant) gene segments. y), J and C g gene segments. g H chains have V,, DH ((diversity), Antigenic determinants on immunoglobulins mouse IgG1 mouse IgM "What makes human IgG1 different f from human h IgM." I M" a. Isotypic determinants mouse IgG1 strain A mouse IgG1 strain B "What makes my IgG1 different from yours." b. Allotypic determinants "What What makes my anti-tetanus different from my anti-pertussis" mouse IgG1 against Ag a mouse IgG1 against Ag b c. Idiotypic determinants During g B cell development p in the bone marrow,, the gene g segments undergo gene rearrangement by bringing together one V, (one D), and one J gene for joining to a C gene segment, a process known as somatic recombination. This process involves excision and splicing of gene segments. The DNA-splicing DNA splicing enzymes are encoded by recombination activator genes, RAG-1 and RAG-2. RAG knock-out mice has no functional T and B cells. H chain gene rearrangement occurs early in B cell development at the Pro-B cell stage (before κ chain gene rearrangement). κ chain h i gene rearrangementt occurs before b f λ chain h i rearrangement at the immature B cell stage in human and mice. A unique recombination occurs in each B cell • each B cell combines these gene segments to make an Ab chain like shuffling a deck of cards - V, D, and J for the heavy chain, V and d J ffor th the li light ht chain h i • since i there th are multiple lti l types t off eachh gene segment, there are many thousands of possible V-D-J combinations so that each B cell gets a unique combination of segments! Unique combination of segments becomes joined by somatic gene rearrangement Combinatorial Diversity Human κ chain gene: (40 Vκ) (5 Jκ) = 200 possible distinct κ V regions. Human λ chain gene: (30 Vλ) (4 Jλ) = 120 possible distinct λ V regions. Total possible L chain V regions: 200 + 120 = 320 Human H chain gene: (40 VH) (25 DH) (6 JH) = 6,000 possible distinct H V regions. (Each of the VH region can join to any one of the C gene segments) Each of the 320 different L chains could combine with each of the , possible p heavyy chains. 6,000 (6,000 H) (320 L) = 1.92 x 106 different ab specificities. Junctional and Insertional Diversity • When the 3' end of V and 5' end of J join together together, the point of joining does not always need to be the same. In this way, the nucleotide triplet which encodes one a.a. may be different each time the same V and J segments are joined. • Similarly in H chains, additional variability occurs at the D-J and V-D-J joining points. • In addition to imprecise joining, additional nucleotides may also be inserted at the junction during rearrangement rearrangement. The inserted nucleotides are found in the CDR3 of both H and L chains, and are called P P- or N N-nucleotides. nucleotides. P-nucleotides are template-based. N-nucleotides are nontemplate-based, i.e. not encoded in the gene segment. The endpoint is additional nucleotides and therefore new amino acid sequences are generated in CDR3 CDR3. Somatic hypermutation yp After boosting immunization, point mutations may occur in the CDRs. Thi diversity This di it is i created t d as a result lt off point i t mutation t ti iin certain "hot spots" and less-defined secondary structural features in the V region. region Junctional Diversity and Somatic Hypermutation together increases the B cell repertoire to 10 11 different specificities. Allelic Exclusion Chromosomes are diploidy. One H chain will undergo rearrangement. If completed, rearrangement of the other (allele) will be prohibited. prohibited If it fails, the other (allele) will continue. The same for L chains. The end result is that each B cell will use only one parental chromosome. This phenomenon is known as allelic exclusion. This is why no B cell will express both κ and λ L chain. Transcription p and Class Switching g Transcription can stop at 3' end of μ gene segment. This yields VDJCμ chain (μ chain). Alt Alternatively, ti l ttranscription i ti can go th through h th the Cμ and d stops t after the δ gene segment. The intervening Cμ will then be spliced out and yield a VDJCδ chain (δ chain). chain) This occurs at mature B cell stage of development. Therefore, mature B cells can express both IgM and IgD sharing the same VHDHJH and the same VJ of the L (κ or λ) chain. This means that the same Ag specificity of different isotypes can be found on a given mature t B cell. ll A single B cell may produce Ig of different isotypes but it always produces an Ig of only ONE antigenic specificity specificity. Class (Isotype) Switching B cells ll mature t iin b bone marrow express b both th IIgM M and d IIgD. D They then join the blood and recirculate in the periphery. In the periphery, periphery B cells can potentially switch isotypes by producing sequentially IgM→IgG →IgA →IgE. The switching of isotype is mediated by the Switch (S) region in the introns 5' to each of the C gene segment ( (except t for f δ). δ) Therefore, the same specificity can be expressed as distinct Therefore isotypes starting always with IgM. Class switching is largely regulated by cytokines (IFN-γ for IgG2 class, and IL-4 for IgG1 and IgE) produced upon Ag stimulation. ti l ti