* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Possible Ligand-binding Proteins in the Olfactory Epithelium of the
Silencer (genetics) wikipedia , lookup
Genetic code wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Metalloprotein wikipedia , lookup
Gene expression wikipedia , lookup
Biochemical cascade wikipedia , lookup
Point mutation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Expression vector wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Paracrine signalling wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Homology modeling wikipedia , lookup
Interactome wikipedia , lookup
Biochemistry wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein purification wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Signal transduction wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
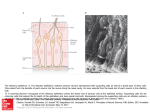
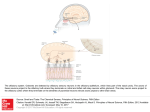
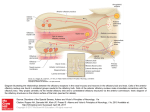
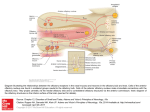
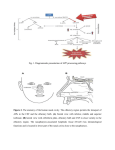
S-1-1 Possible Ligand-binding Proteins in the Olfactory Epithelium of the Japanese Common Newt Taro Mizumoto1, Jiro Muroran2 and Saburo Iburi 1 Department of Materials Science and Engineering, Muroran Institute of Technology, 27-1 Mizumoto, Muroran 050-8585, Japan 2 Applied Physics and Chemistry, The University of Electro-Communications, Chofugaoka 1-5-1, Chofu, Tokyo 182-8585, Japan Email: [email protected] 1. Introduction. Recently, the number of the chemicals has increased tremendously in our environment. Some of these chemicals caused harmful effect to living organisms including humans. The mechanism causing such toxic effects on the organisms are still not well-understood and possibly different from the each chemical. However, the first step of the toxic effects should be an interaction between such environmental chemicals and molecules such as proteins or other biological molecules of living organisms. The large part of these proteins are embedded in the cytoplasmic membranes of cells building living organisms and called “membrane receptors”. However, in the case of dioxin, it is very hydrophobic, easily passes the cytoplasmic membrane, and binds aryl-hydrocarbon receptor (AhR) in the cell. The complex of dioxin and AhR cause the change of gene expression pattern of the cell, which results in harmful effects on organism. Therefore, to understand the mechanism of toxic effects of a chemical it is very important to understand the interaction between the chemical and the target protein, receptor. For the model system to study this interaction, an olfactory system of animal is very suitable. We have been studying the olfactory system of the newt and found a protein (Cp-lipocalin) that shows amino acid sequence homology to AhR. 2. Results and Discussion. 2.1 Several types of possible ligand-binding protein s were found in a cDNA library of the olfactory epithelium of the newt. We constructed a cDNA Library with messenger RNA obtained from the olfactory epithelium of the newt, Cynopus pyrrhogaster. By a preliminary exhaustive analysis of the library, we found three types of clones that are abundantly expressed in the olfactory epithelium of the newt. The deduced amino acid sequence of two of them, clone #75 and #89, showed high sequence homology to each other and to the members of lipocalin super-family, frog olfactory specific protein (OSP) and Hyla japonica lipocalin-type protein, respectively. They were tentatively named as Cp-OSP and Cplipocalin. Lipocalins are a large family of small proteins that are characterized by the ability to bind small hydrophobic molecules. Their primary sequences are diverged, but the 3D structure of the lipocalins shows a conserved folding pattern. The molecular phylogenetic tree constructed by the neighbor-joining methods confirmed that Cp-OSP and Cp-lipocalin are highly similar each other and they belong to the lipocalin super-family (Fig. 1). Another class of the possible lignad-binding protein was also found in the olfactry epithelium of the newt. Analysis of the nucleotide sequence of the third clones revealed a high sequence similarity to rat potential ligand-binding protein (RY2G5; PLBP). The deduced amino acid sequence showed homology to rat ligand-binding protein and also to human BPI (BPI : Bactericidal/PermeabilityIncreasing protein) family proteins. Therefore, it was named as Cp-PLBP. 0.487 Gallus gallus CALⅡ 5 0.118 9 0.487 Homo sapiens MSFL2541 5 0.014 1 0.099 5 0.162 8 0.100 9 0.433 4 [GENETYX-MAC:Evolutionary Tree] Method:UPGMA 0.443 6 No 75 : Cp-OSP 0.443 6 No. 89 : Cp-lipocalin 0.086 Hyla japonica lipocalin 2 0.086 Bufo marinus lipocalin 2 0.187 Xenopus laevis cpl-1 1 0.720 Rana nigromaculata osp 0 Fig. 1. Molecular phylogenetic tree of Cp-OSP and Cp-lipocalin constructed with Genetyx Mac UPGMA program. 2.2 Cp-OSP and Cp-lipocalin were expressed in olfactory epithelium. To elucidate tissue specificity of Cp-OSP and Cp-lipocalin, total RNA was isolated from several tissues of the newt and the northern blotting analysis was performed with alkaline phosphatase-labeled anti-sense probe of Cp-OSP or Cp-lipocalin. The results showed that the transcripts of the two clones were highly expressed in the newt olfactory epithelium but not in brain, liver and intestine. 2.3 The transcripts of the three possible ligand-binding proteins were expressed in Bowman s gland. The digoxygenin-labeled anti-sense riboprobe of each three possible lignad-binding proteins was made and in situ hybridization was performed with a frozen section of the newt olfactory epithelium. Based on the analysis, they were all expressed in cells composed of the Bowman’s grand, which secrete mucus covering the olfactory epithelium. The results suggest that these possible ligandbinding proteins are produced in the Bowman’s grand and transferred to the mucus layer. The finding that the deduced amino acid sequences of Cp-OSP and Cp-lipocalin have a possible signal sequence in their N-terminal region is not contradictory to the suggestion. In the case of Cp-PLBP, because the full length cDNA has not been unfortunately isolated yet, it is not sure that Cp-PLBP is really secreted protein. 2.4 The distribution of the cells which express each possible ligand-binding protein showed different pattern on the olfactory organ of the newt. From the results of in situ hybridization with a frozen section of the olfactory epithelium, it is not elucidated whether these proteins are produced in the same cells composed Bowman’s grand. To clarify the question, we used serial frozen sections of the nasal cavity of C. pyrrohogaster, and studied whether these proteins are produced in the same cells. The results of the in situ hybridization analysis of the whole nasal cavity of the newt showed that the cells which express each possible ligand-binding protein are different in their distribution pattern on the olfactory organ. Although the transcripts of Cp-OSP were observed in almost all Bowman’s grand of the nasal cavity, those of Cp-lipocalin were observed only in Bowman’s grand on central and dorsal part of the nasal cavity. The Bowman’s grand on the ventral part of the cavity did not show the signal. The cells which express Cp-PLBP showed a different distribution pattern from that of lipocalin-family proteins; only the Bowman’s grand on the central part of the cavity expresses Cp-PLBP. 3. Concluding remarks. Our results showed that the different types of possible ligand-binding proteins are expressed in the different parts of the nasal organ of the newt and that possibly form heterogeneous mucous environment on olfactory epithelium. Although the physiological role of the proteins and their importance in olfactory sensation are remained unclear, their possibility of binding small molecules, such as odorant, and the difference in distribution on the mucus layer suggest that a pre-discrimination of odorant is performed by these proteins before odorants reach to olfactory receptors.