* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download this lesson
Genome evolution wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Western blot wikipedia , lookup
Molecular cloning wikipedia , lookup
DNA sequencing wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Whole genome sequencing wikipedia , lookup
DNA supercoil wikipedia , lookup
Molecular evolution wikipedia , lookup
Size-exclusion chromatography wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Non-coding DNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
SNP genotyping wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Genomic library wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Agarose gel electrophoresis wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
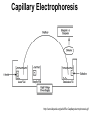
Capillary Electrophoresis and the Human Genome Sequencing the Human Genome • Medical uses – Genetic predisposition to diseases • Forensic uses – Matching DNA samples • Potential for “personalized medicine” – Testing for genetic abnormalities and customizing treatment The Sequencing Challenge • 1990: DNA and RNA sequencing used Sanger Method: 1. 2. 3. 4. Amplification – using E. coli Labeling – using radioactive ddntps Separation – using electrophoresis Reading – done manually 60 years would be needed to sequence the 3 billion base-pairs in human DNA Sanger Method • Advantages: – Automation to a certain degree • Disadvantages: – Time: 60 years would be needed to sequence the 3 billion base-pairs in human DNA Electrophoresis • Applied direct current to separate molecules based on charge and size • The higher the voltage, the faster separation will be achieved • Can be done in liquid or gel medium, slab or capillary Voltage and Speed of Separation • The migration rate v of an ion in cm/s is given by: v = μE • A higher electric field (E), achieved through a higher applied voltage, will result in a faster separation • μ can be changed only slightly using different conditions like pH, surfactants and buffers Speed verses Accuracy • The gel below exhibits “joule heating”, distortions caused when the voltage applied is too high for the conditions used Protein products from E. coli Photo: Katie Kollitz 2008 Sample 96 Lane Gel Electrophoresis Capillary Electrophoresis • The surface area of a capillary is very high compared to its volume, so joule heating disappeared • The narrow capillary has a high resistivity, meaning current will stay low even at high voltages • Voltages are much higher in CE Capillary Electrophoresis http://en.wikipedia.org/wiki/File:Capillaryelectrophoresis.gif Inside the Capillary • Electric Field provides separative power, causes bulk flow • Electrophoretic Mobility: property of each analyte • Electroosmotic Flow: allows for elution and separaton of positive, neutral and negative molecules Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ Na+ http://content.answers.com/main/content/img/McGrawHill/Encyclopedia/images/CE226400FG0010.gif Advantages • The small size of a capillary offers high resistance, so the electric field can be large while keeping current low, speeding separations and improving resolution • A smaller sample size may be used • Because the samples elute from one end, quantitative detectors may be used • Electroosmotic flow allows for separation of negative, positive and uncharged molecules Capillary Gel Electrophoresis • Since DNA has a uniform charge to size ratio, a gel must be used to introduce frictional forces • CGE retains the advantages of speed, small sample size and quantitative output Raw Results Capillary gel electrophoresis of a DNA sequence using fluorescently tagged primer & ddCTP: spikes in voltage indiate the presence of a Cytosine residue Four Color Fluoresence • Four color fluorescence allowed for data to be read by a machine instead of manually Future Developments • Concept of lab on a chip – Preparation step is the only one that hasn’t been automated – Lab on a chip eliminates amplification step and separation step – Labeling and reading happen simultaneously – Requires intense computational ability True Signal Molecule Sequence by Helicos BioSciences • Two flow cells filled with billions of copies of sample DNA attached to surface. • DNA polymerase catayzes reaction using one added fluorescently tagged ddNT. • Wash out free nucleotides and position of ddNTs recorded. • Remove fleorescently tagged group leaving behind generated complementary strands • Repeat for other bases Results • Multiple four base cycles provide 25 base length sequences! Fluoresecence of 1 and 2 in cycle X indicate the presence of a G nucleotide