* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download NUCLEOTIDES METABOLISM Nucleotide
Oligonucleotide synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Microbial metabolism wikipedia , lookup
Genetic code wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Metalloprotein wikipedia , lookup
Peptide synthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
15-Hydroxyeicosatetraenoic acid wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Butyric acid wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
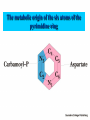
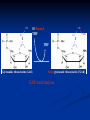
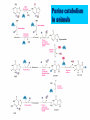
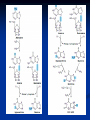
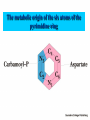
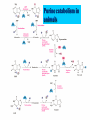
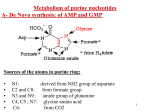
NUCLEOTIDES METABOLISM Nucleotide biosynthesis: - Most organisms can make purine and pyrimidine nucleotides via de novo (from scratch) pathways - They can also recover nucleotides from diet - Rapidly dividing cells require large amounts of RNA and DNA - In these cells, large quantities of nucleotides are needed - These pathways are attracting targets for treatment of cancer and infectious microorganisms - Many antibiotics and anticancer drugs are inhibitors of nucleotide biosynthesis Nucleotide biosynthesis: Principles and differences - Purines: Successive addition of atoms Ribose-5-phosphate serves as a base for the addition of successive atoms derived from common metabolic intermediates - Pyrimidines: Synthesized directly from two common metabolic intermediates Nucleotides are synthesized prior to their linkage to ribose-5-phosphate The metabolic origin of the nine atoms in the purine ring system The metabolic origin of the six atoms of the pyrimidine ring Inosine-5'-P Biosynthesis The purine ring is built on a ribose-5-P foundation First step: ribose-5-P must be activated - by PPi 5-phosphoribosyl-a-pyrophosphate (PRPP) is limiting substance for purine synthesis But PRPP is a branch point so the next step is the committed step - Gln PRPP amidotransferase Azaserine - Gln analog - inhibitor/anti-tumor Steps in Purine biosynthesis 1- Activation of R5-P and formation PRPP %-phosphoribose pyrophosphation, 2- Addition of N2 from Glutamine 3- Condensation with glycine (need ATP) 4- Addition of Carboxyl group C=O from N10 THFA ( Tetra hydro folic acid) 5- Addition of N2 from glutamine (need ATP) 6- Ring closure need ATP ,removal of water molecule PRPP: A Central Metabolite in De Novo and Salvage Pathways 5-Phospho-a-D-ribosyl-1-pyrophosphate (PRPP) is an activated ribose-5-phosphate derivative used in both salvage and de novo pathways. PRPP synthetase Phosphoribosyltransferase (HGPRT) Gln Glu, PPi PRPP amidotransferase AMP, GMP Gly, ATP GAR synthetase ADP, Pi NH2 H2C C O Glycinamide ribonucleotide (GAR) NH2 H2C NH2 10-FormylTHF C H2C C THF O CHO O Glycinamide ribonucleotide (GAR) Formylglycinamide ribonucleotide (FGAR) GAR transformylase NH2 H2C C O NH2 CHO Gln, ATP Formylglycinamide ribonucleotide (FGAR) H2C Glu, ADP, Pi CHO C HN Formylglycinamidine ribonucleotide (FGAM) FGAR amidotransferase 6- Ring closure by dehydration removal H2O (need ATP Mg 7- Addition of Carboxyl group from N10THFA 8- Addition of Aspatate ( for addition N2 ) 9- Removal fumarate 10 –Addition Carboxyl group from N10 THFA 11- Ring closure (remove water molecule) and formation IMP The synthesis of AMP and GMP from IMP Making AMP and GMP Reciprocal control occurs in two ways GTP is the energy input for AMP synthesis, whereas ATP is the energy input for GMP AMP is made by N addition from aspartate GMP is made by oxidation at C-2, followed by replacement of the O by N (from Gln) Potent inhibitors of purine nucleotide synthesis -- structural analogs of glutamine -- glutamine amidotransferases Nucleotides are active in metabolism primarily as the nucleoside triphosphates. GMP and AMP are converted to their corresponding triphosphates through two successive phosphorylation reactions. Conversion to the diphosphates involves specific ATP-dependent kinases. Guanylate kinase Adenylate kinase GMP + ATP GDP + ADP AMP + ATP 2ADP Purine degradation and clinical disorders of purine metabolism Formation uric acid All purine nucleotide catabolism yields uric acid. Purine catabolism in primates ends with uric acid, which is excreted. Most other animals further oxidize the purine ring, to allantoin and then to allantoic acid, which is either excreted or further catabolized to urea or ammonia. Regulation of purine biosynthesis Purine Degradation Purine catabolism leads to uric acid Nucleotidases and nucleosidases release ribose and phosphates and leave free bases Xanthine oxidase and guanine deaminase route everything to xanthine Xanthine oxidase converts xanthine to uric acid Xanthine oxidase can oxidize two different sites on the purine ring system Purine Degradation Purine catabolism leads to uric acid Nucleotidases and nucleosidases release ribose and phosphates and leave free bases Xanthine oxidase and guanine deaminase route everything to xanthine Xanthine oxidase converts xanthine to uric acid Xanthine oxidase can oxidize two different sites on the purine ring system Purine catabolism in animals PNP: Purine nucleoside phosphorylase ADA: Adenosine deaminase (Muscle) Nucleotidase Nucleotidase ADA PNP PNP Guanine deaminase Xanthine oxidase Hypoxanthine Xanthine oxidase Xanthine Uric acid Figure 22.7: Catabolism of purine nucleotides to uric acid. Xanthine Oxidase and Gout XO in liver, intestines (and milk) can oxidize hypoxanthine (twice) to uric acid Humans and other primates excrete uric acid in the urine, but most N goes out as urea Birds, reptiles and insects excrete uric acid and for them it is the major nitrogen excretory compound Gout occurs from accumulation of uric acid crystals in the extremities Allopurinol, which inhibits XO, is a treatment Pyrimidine nucleotide metabolism Pyrimidine nucleotide synthesis occurs primarily at the free base level, with conversion to a nucleotide occurring later in the unbranched pathway. Pyrimidine synthesis begins with formation of carbamoyl phosphate. In enteric bacteria, this enzyme represents an example of feedback control. The enzyme is inhibited by the end product CTP and activated by ATP. Pyrimidine Biosynthesis In contrast to purines, pyrimidines are not synthesized as nucleotides Rather, the pyrimidine ring is completed before a ribose-5-P is added Carbamoyl-P and aspartate are the precursors of the six atoms of the pyrimidine ring CPS II Carbamoyl phosphate for pyrimidine synthesis is made by carbamoyl phosphate synthetase II (CPS II) This is a cytosolic enzyme (whereas CPS I is mitochondrial and used for the urea cycle) Substrates are HCO3-, glutamine, 2 ATP The metabolic origin of the six atoms of the pyrimidine ring Carbamoyl phosphate synthetase II reaction 1- CO2 + GLn + 2 ATP + H2O= Carbamoyl phosphate 2- Addition of Asp acid =Carbamoyl aspatic acid 3- Ring closure (dehydration) by action of enzyme oorotase = Di hydro oorotic acid 4- Di-hydro –oorotic acid → Oorotic acid By the action of the enzyme Di-hydro –oorotic acid DH 5- OA + PRPP = OMP ( oorotate phosphoribosyl Transferase enzyme) 6- OMP → UMP (Enz= orotidylic acid decarboylase ,Process= Decarboxylation CTP synthesis from UTP Purine catabolism in animals Control of pyrimidine biosynthesis in bacteria and animals Figure 22.11: Catabolic pathways in pyrimidine nucleotide metabolism.