* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Homologs: behave independently in mitosis Tfm: secondary and
Nutriepigenomics wikipedia , lookup
Gene desert wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genome evolution wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Minimal genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Copy-number variation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Designer baby wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Gene expression programming wikipedia , lookup
Genomic imprinting wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Microevolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
Y chromosome wikipedia , lookup
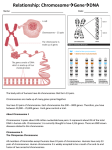
Homologs: behave independently in mitosis Tfm: secondary and tertiary sex determination fail as a male Anticipation: Increase in severity from one generation to the next 1:2:1: ratio of 2 strand: 3 strand: 4-strand double crossovers Sister chromatid adhesion: lost at start of Anaphase I Sister chromatids: adhere to one another in prophase I 10 map units: interference = 1 PD=NPD: genes on non-homologous chromosomes Essential genes: about 1/3 of all genes 5-bromouracil: a base analog trait rare: assume unrelated individuals homozygous or hemizygous for normal allele auxotroph: requires nutritional supplementation beyond that required by wild type SNP: due to a base pair substitution Addition rule: can be used when events are mutually exclusive Zygotene: synaptinemal complex forms Chromatid interference: does not exist Indel: e.g. deletion of a few base pairs Bivalent: physical association of two homologs 3 strand doubles: produce T asci Z: one copy present in female Interphase: DNA replication occurs in some, but not all, cases Pleiotropic: one allele causes more than one phenotype Law of Segregation: each gamete receives only one copy of each gene Reductional division: each chromosome has 2 chromatids after division completed, each gamete receives only one copy of each gene (Meiosis I) Germ cell: cell that gives rise to gametes Repulsion: alleles on different homologs Chaisma frequency = 20%: recombination frequency = 10% Chi-squared = sum (observed-expected)2/(expected) Central element: appears progressively during synapsis P value = 6%: data not significantly different from expectation Adjacent I: homologous centromeres segregate to opposite poles Chromatid interference: level determines fraction of double crossovers that are 2-strand, 3-strand, and 4-strand doubles Expressivity: degree to which phenotype is altered in affected individuals Mendel’s 1st law: the law of segregation: each trait is controlled by particulate factors that occur in pairs; during gamete formation, the members of a pair separate from each other so that each gamete receives only one; the double number is restored upon fertilization Mendel’s 2nd law: the law of independent assortment: the segregation of each gene pair during gamete formation is independent of all others Metacentric: centromere near center of chromosome Sub-metacentric chromosome: centromere more near end Acrocentric chromosome: centromere almost at very end Telocentric chromosome: centromere pretty much at end Karyotype: chromosome sets as seen at end of prophase Matroclinous: daughter is like mother in phenotype even in criss-cross inheritance (result from nondisjunction in anaphase) Patroclinous: son is like father in phenotype because of nondisjunction Nondisjunction: homologs fail to let go (dragged to one pole) or homologs never pair together (no basis for segregation) In Drosophila, sex determined by number of X chromosomes; Y chromosome confers male fertility (XXX and YO die, XX female, XO and XY male) Hemizygous: present in only one copy (in male, such as XY) Moths, birds, and butterflies: ZW female, ZZ male Mammals: XO fertile female (Turner’s syndrome), XXY sterile male (Klinefelter’s Syndrome), XXX female Y chromosome determines maleness in mammals Testicular Feminization Gene (Tfm): primary: gonad testes, secondary: fails female genitalis, tertiary: fails Testes determining factor (TDF) and Sex determining region on Y (Sry) Propositus: individual that brought attention of pedigree to geneticists (indicated by arrow) When alleles are together on same homolog, they are said to be in cis with one another or in coupling. When alleles are on different homologs, they are said to be in trans or in repulsion. Linkage: association of genes on same chromosome, genes said to be linked Recombination: production of new gamete types Recombination frequency: nonparental gametes/total gametes * 100% Prophase I: leptotene (thin thread), zygotene (paired thread, beads on string), packytene (thick thread), diplotene (2 thread) Synapsis: intimate and precise pairing of homologs occurs in zygotene and the complex is seen in packytene Locust: position of gene on chromosome (plural, luci) 1% rec. = 1 map unit (centimorgan) undetected double crossovers lead to an underestimate of distance between genes double crossovers are rarer than expected based on assumption of independence of crossing over in adjacent intervals Interference: occurrence of one crossover inhibits others from occurring nearby Coefficient of coincidence: (observed # doubles)/(expected # doubles) (between 0 and 1) Interference: 1- coefficient of coincidence Interference = 1 within 10 map units, there are no doubles within 10 map units 3 types of tetrads: parental ditype (PD), tetratype (T), nonparental ditype (NPD) NPD<<PD genes are linked If not linked, PD=NPD because of Mendel’s second law Chromatid interference: influence of chromatids selected for 1st crossover on chromatids selected for second DOES NOT EXIST Chiasma interference: real, inhibition by one crossover of others occurring nearby Even if a double crossover occurred 100% of the time between a and c, rec. freq. would equal 50% because of no chromatid interference Indel: insertions of deletions of a few base pairs Polymorphism: presence of more than one common form in a population Founder effect: alteration in allele frequency due to the founding of the population by a small # of individuals