* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
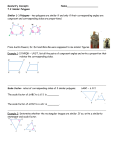
Download Name______KEY Genetics C3032 - Examination #2
Skewed X-inactivation wikipedia , lookup
Koinophilia wikipedia , lookup
Gene desert wikipedia , lookup
Frameshift mutation wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Essential gene wikipedia , lookup
Genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Y chromosome wikipedia , lookup
Neocentromere wikipedia , lookup
Nutriepigenomics wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Oncogenomics wikipedia , lookup
Minimal genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Pathogenomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
X-inactivation wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression profiling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup






![EM5_09[1] - Graduate Institute of International and Development](http://s1.studyres.com/store/data/008685416_1-fecdc75f61e8e66f8a0f9ffbf33b86ae-150x150.png)