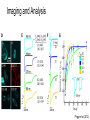
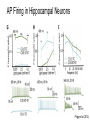
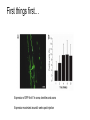
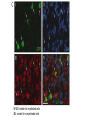
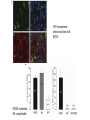
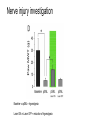
* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Optogenetics - FSU Program in Neuroscience
Neural engineering wikipedia , lookup
Development of the nervous system wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Multielectrode array wikipedia , lookup
Neuroanatomy wikipedia , lookup
Single-unit recording wikipedia , lookup
Stimulus (physiology) wikipedia , lookup
Microneurography wikipedia , lookup
Feature detection (nervous system) wikipedia , lookup
Neuroregeneration wikipedia , lookup
Electrophysiology wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Optogenetics Genevieve Bell Daniel Blackman Brandon Chelette Membrane Biophysics - Fall 2014 Optogenetics • Integration of optics and genetics that allows for experimental control of events within a specific cell Before Optogenetics • The spatial and temporal resolution of modulation in the brain left much to be desired • Spatial resolution: stimulation with electrodes does not distinguish between different cell types (Francis Crick, 1979) • Temporal resolution: multi-component systems require a long time to occur • Insufficient for millisecond time scales needed to analyze cellular events such as action potentials Before Optogenetics • It was known that there were single component, light activated ion pumps in bacteria • Vast array of microbial opsin genes Optogenetics – the basic concept • Express a light-activated ion channel (isolated from bacteria) in a specific sub-population of neurons • Illuminate neurons to modulate activity • Modulation depends on type of channel • Record output (e-phys, fMRI, behavioral, etc.) Initial Problems • It was presumed that: • Microbial proteins would not be expressed very well in mammalian cells • Photocurrents not strong enough to control neurons • Retinal (co-factor) would have to be added to the cell type of interest Initial Problems • It was presumed that: • Microbial proteins would not be expressed very well in mammalian cells (bacterial opsin expressed reliably in mammalian neurons. Boyden 2005) • Photocurrents not strong enough to control neurons • Retinal, a requisite cofactor for opsins, would have to be added to the cell type of interest Initial Problems • It was presumed that: • Microbial proteins would not be expressed very well in mammalian cells (bacterial opsin expressed reliably in mammalian neurons. Boyden 2005) • Photocurrents not strong enough to control neurons (bacterial opsin provided sustained control of action potential. Boyden 2005) • Retinal, a requisite cofactor for opsins, would have to be added to the cell type of interest Initial Problems • It was presumed that: • Microbial proteins would not be expressed very well in mammalian cells (bacterial opsin expressed reliably in mammalian neurons. Boyden 2005) • Photocurrents not strong enough to control neurons (bacterial opsin provided sustained control of action potential. Boyden 2005) • Retinal, a requisite cofactor for opsins, would have to be added to the cell type of interest (all vertebrate tissues contain sufficient all-trans retinal. Douglass 2008) Optogenetics • Allows for high resolution temporal and spatial modulation of the activity of a cell • The population of neurons being targeted, the timing of the modulation, and the nature of the modulation are all under experimental control Optogenetics: a diverse toolkit • The optogenetic effect experienced by a cell will depend on many factors • • • • The properties of single-component opsin being used The efficiency of the expression of that opsin The source/wavelength/intensity of the light The location and density of the population of neurons being investigated Optogenetics: a diverse toolkit • Wide variety of opsins Optogenetics: a diverse toolkit Four main categories of opsins: • Fast excitation • Fast inhibition • Step function • Biochemical modulation Optogenetics: a diverse toolkit Biochemical modulation • Opsin + G-protein coupled receptor • Activated by light, but not an ion channel • Illumination leads to intracellular signaling cascades • Can be considered slow excitatory or slow inhibitory based on the nature of the G protein signaling pathway • Provide precise control of intracellular signals (cAMP, GTPase, etc.) Optogenetics: a diverse toolkit Fast excitation • Channelrhodopsins • Cation channels • Modified via mutagenesis with optimization in mind, but can result in unanticipated or undesired effects • Gain-of-function tool Optogenetics: a diverse toolkit Fast excitation Optogenetics: a diverse toolkit Fast inhibition • Chloride & proton pumps • Also engineered for optimal function • Loss-of-function tool Optogenetics: a diverse toolkit Fast inhibition Optogenetics: a diverse toolkit Step function opsins (SFOs) • Exhibit bistability • Much slower deactivation rate • Product of molecular engineering • Cation channels • Larger disparity between activating wavelength and deactivating wavelength Optogenetics: a diverse toolkit Step function opsins (SFOs) Optogenetics – a diverse toolkit • Vast array of opsins available to be used as a result of attempted optimization • Point mutations • Codon insertion/deletion/replacement • Small changes can lead to beneficial or detrimental changes in: • Current generation • Opening/closing kinetics • Sensitization/Desensitization Optogenetics – targeting your opsin • Viral injection • LV , AAV • Projection targeting • Dendrites or axons • Transgenic targeting • Transgenic mouse lines that are not under recombinase-dependent control • Spatiotemporal targeting • Birthdate of cells , specific layer Optogenetics – light delivery • Assuming you are expressing the correct opsin in the desired cell population, you now need to somehow get light to those cells • There are several facets to consider and the best choice will depend on your experiment • • • • Excitation vs inhibition vs bistable Wavelength Intensity Duration Optogenetics – light delivery • Brain tissue scatters and absorbs light • Different tissues scatter and absorb light in different ways (Ex: myelinated tissue scatters light significantly) Optogenetics – light delivery Different wavelengths of light penetrate brain tissue better than others Optogenetics – light delivery • Surface targets: cell culture, brain slices, etc • Easily accessible, spot illumination • Deep Targets: in vivo targets that are not on the outer surface of the brain • Minimize damage • Fiber optics Optogenetics – light sources • Variety of light sources: • Lasers • LEDs • Incandescent • Depending on where you need to deliver your light and how, you need to select the correct light Optogenetics – the good • Significant experimental control • High resolution temporal and spatial control • Specific cell type population • Millisecond control on time scale of cellular events like action potentials • Wide variety of opsins available • Can be applied to more fields than just neuroscience (cardiac muscle, skeletal muscle, etc) • Potential disease models Optogenetics – the bad • Damage to surrounding tissue (depending on location of target) • Light absorption heat damage tissue or affect physiological events • Viral infection and/or expression of exogenous proteins can lead to unwanted alterations in cell capacitance, physiological activity, structural abnormalities, and toxicity • Second and third degree currents can confound actual effects • Causality can not always be proven The ugly take home message • Optogenetics is a burgeoning technique that has provided neuroscientists with a much more finely-tuned way to manipulate the cellular activities of a specific population of cells they are interested in. But with any relatively new technique, there are still kinks that need to worked out Color-tuned Channelrhodopsins for Multiwavelength Optogenetics Matthias Prigge, Franziska Schneider, Satoshi P. Tsunoda, Carrie Shilyansky, Jonas Wietek, Karl Deisseroth, and Peter Hegemann A presentation by Daniel Blackman Modifying ChRs to Overcome Limitations • Limitations of ChR2s 1. 2. 3. 4. 5. Low expression Small conductance Inappropriate kinetics Partial inactivation Ion selectivity • Retinal pocket changes affect absorption, kinetics, and membrane targeting 1. 2. 3. 4. Glu123Thr/Ala (ChETA variants) Cys128Ser or Asp156Ala Glu90, Glu123, Leu132, or His134 C1 and C2 helix swapping What Previous Studies Have Taught Us • Volvox C1 (V1) absorption max • Pyramidal neurons • Dual-color activation • Spectrally separated absorption • Large photocurrents • Different operational sensitivities Spanning the Spectrum • Operational Light Sensitivity • Calcium indicators or voltage sensors • Molecular engineering • Helix Swapping • Global rearrangements Global Structural Rearrangements Prigge et al (2012) Imaging and Analysis Prigge et al (2012) Fast and Slow-Cycling Prigge et al (2012) AP Firing in Hippocampal Neurons Prigge et al (2012) Dual Light Excitation and Ion Selectivity Prigge et al (2012) Current-voltage relationships Prigge et al (2012) Conclusion • Identified helices H6 & H7 responsible for absorption differences, and helices H1 & H7 for membrane integration • Achieved independent activation of distinct neural populations through AA mutations: • • • • ChR2 with absorption maxima 461-492 nm (blue) C1V1 chimeric variant with absorption maxima 526-545 nm (green) C1V1-B & C2-LC-TC provide better expression & lower inactivation Further color tuning possible through exploring further AA mutations Fast-conducting mechanoreceptors contribute to withdrawal behavior in normal and nerve injured rats Ririe DG, et al. 2014. Pain. Membrane Biophysics – Fall 2014 Background - Pain • Transduction of noxious (or potentially noxious) stimuli from periphery to central nervous system • Complex integration with emotional and cognitive signals perception of pain Background - Pain A-fibers – “first pain” C-fibers – “second pain” Julius & Basbaum 2001 Background - Pain Ablation of C-fibers does not eliminate pain behavior Other afferents might be responsible for at least some portion of ongoing pain Background - Experiment • Neurons of interest: fast-conducting, myelinated, nociceptive, high-threshold mechanoreceptors = AHTMRs • Investigate: what is the role of these neurons in pain signal transduction under normal conditions and under neuropathic conditions? Background - Experiment • Selectively inhibit these specific neurons (AMHTRs) using expression of light-activated proton pump • Male Sprague-Dawley rats: intrathecal injection of AAV8 with CAG/ArchT/GFP tag Light activates proton pump Pumps protons intra extra Hyperpolarizes cell / Reduce excitability / Inhibit activity First things first… Expression of GFP-ArchT in soma, dendrites and axons Expression maximized around 4 weeks post-injection NF200: marker for myelinated cells IB4: marker for unmyelinated cells GFP co-expressed almost exclusively with NF200 NF200: myelinated IB4: unmyelinated In vitro recordings • Excised DRG four weeks after injection • Single-electrode, continuous-current clamp recording of GFP positive cells • 0.1 nA – 4.0 nA in 0.1 nA increments until an AP was evoked to determine threshold In vitro recordings Light exposure increased threshold Light exposure hyperpolarized cells In vivo recordings • 3 – 8 weeks after injection • L4 DRG ganglia exposed • Receptive field, conduction velocity, and electrophysiological profile determined In vivo recordings In vivo recordings Only AHTMRs affected by light LTMR: myelinated low threshold mechanoreceptors AHTMR: myelinated high-threshold mechanoreceptors CHTMR: unmyelinated high-threshold mechanoreceptors In vivo recordings AMHTR recordings: inhibition of AP when soma exposed to light In vivo recordings Stimulus at receptive field in paw results in AP AP eliminated upon illumination of DRG In vivo recordings (transcutaneous) Same idea: still recording from DRG, but light is now targeted at receptive field in paw In vivo recordings (transcutaneous) AMHTR recording: transcutaneous light still inhibits AP firing In vivo recordings (transcutaneous) In vivo recordings (transcutaneous) Dose-response curve So far, so good • Intrathecal injection of viral vector with GFP-ArchTCAG only expresses in AMHTRs • Expression of GFP-ArchT allows for optical inhibition • In vitro: illumination decreases membrane potential and increases threshold • In vivo: • stimulate soma + illuminate soma = inhibition • stimulate receptive field + illuminate soma = inhibition • stimulate RF + illuminate RF = inhibition • Under “normal” conditions; what happens following a nerve injury? Nerve injury investigation • Following nerve injury, afferents become hyperexcitable • May contribute to chronic pain • Inhibition of AMHTRs may be beneficial for reducing pain after a nerve injury • However, there is no basis to assume that inhibition after a nerve injury will be the same as inhibition in the “normal” state Nerve injury investigation • Induce hyperexcitability via partial ligation of L5 nerve • Investigate the effects of light on hyperexcitability Nerve injury investigation Evidence of hyperexcitability Increased receptive field Nerve injury investigation AMHTR: sham AMHTR: pSNL Nerve injury investigation pSNL: reduction in mechanical threshold and reduction in mechanical withdraw threshold Nerve injury investigation Baseline vs pSNL = hyperalgesia Laser ON vs Laser OFF = reduction of hyperalgesia Nerve injury investigation LTMR AHTMR CHTMR Only AHTMRs inhibited by transcutaneous light on receptive field Nerve injury investigation - summary • pSNL resulted in an increased receptive field, increased sensitivity, reduced mechanical threshold, and reduced withdraw threshold • Optogentic inhibition reduced hyperalgesia • Transcutaneous light only inhibited AMHTRs Conclusions • AMHTRs, which have been established and accepted as “first” pain fibers, seem to play a role in chronic pain as well • Evidenced by correlation between withdraw threshold and AMHTR sensitivity • Also demonstrated behavioral changes under normal conditions and under conditions that simulate pathological nerve damage Stimulating Cardiac Muscle by Light Cardiac Optogenetics by Cell Delivery Zhiheng Jia, MS; Virginijus Valiunas, PhD; Zongju Lu, PhD; Harold Bien, MD, PhD; Huilin Liu, MS; Hong-Zhang Wang, PhD; Barbara Rosati, PhD; Peter R. Brink, PhD; Ira S. Cohen, MD, PhD; Emilia Entcheva, PhD A presentation by Daniel Blackman Tandem Cell Unit (TCU) Zhiheng et al (2011) Proof-of-Principle Cell Delivery System • Cell line using ChR2 plasmid • Whole-cell and dual-patch techniques • Gap-junction uncoupling • Electrical vs optical stimulation Developing and Characterizing the System Zhiheng et al (2011) Validation of the TCU Concept Zhiheng et al (2011) Dual-Patch and Carbenoxolone (CBX) Zhiheng et al (2011) Optical Mapping System Zhiheng et al (2011) Optical Mapping Zhiheng et al (2011) TCU Benefits and In Vivo Consideration • TCU vs Direct: 1. Donor cells (DCs) = nonexcitable and no major repolarization currents 2. DCs have high membrane impedance (small IR currents) 3. Reduced DC-CM vs CM-CM coupling • Optical excitation improvements: 1. Higher membrane impedance DCs 2. Lower membrane impedance CM neighbors 3. Better source-neighbor coupling Conclusion • High spatiotemperal resolution of excitation without affecting essential properties • Potential investigations into optical pacemakers • Five main features of this study: 1. 2. 3. 4. 5. Desired pace rate achieved Finer manipulation of excitation, repolarization, and AP shape In vivo applications (vs viral transfection) Biocompatibility of optic fibers (vs metal electrodes) Improved energy consumption (vs electrical pacemakers)