* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download document
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Designer baby wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Behavioural genetics wikipedia , lookup
Public health genomics wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetic drift wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Medical genetics wikipedia , lookup
Genome-wide association study wikipedia , lookup
Human genetic variation wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
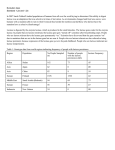
SNPing Lactose By: Mandy Butler, Ying-Tsu Loh and Cheryl Ann Peterson A mother and father and their two young children walk into an ice-cream bar. The two youngsters order delicious 100% whole milk milkshakes, while their parents look on enviously and order non-dairy fruit smoothies. Why do you think the parents didn’t order milkshakes? LACTOSE (pre-assess student understanding) • Lactose, the primary sugar in milk, is hydrolyzed by an enzyme called lactase into the more absorbable monosaccharides, glucose and galactose. • Virtually all humans are born with ability to digest lactose but many lose this ability as they age. • In lactose tolerant individuals, the lactase gene is expressed into adulthood, so eating a milkshake is a pleasant experience. But in people who are lactose intolerant, that lactase gene is switched off, and the consumption of milk products can lead to unpleasant effects. • How is lactose intolerance measured? • What are the symptoms of lactose intolerance? Lactase Survey class on lactose tolerance/intolerance • Show map of lactose tolerance geographically • Ask for ideas about why certain populations are lactose tolerant and others are not • Is there consistency between the map and their condition/experiences? • Introduce concept of phenotype LacIntol-World.png Pre-assess students on SNPs • What might account for this difference in phenotype on a genetic level? • Idea of genetic polymorphisms? – What kinds of polymorphisms are there? – What is a SNP? What evidence is there that lactose tolerance is due to a genetic polymorphism? Which genotype(s) are correlated with lactose tolerance? Enattah et al (2002) Nature Genetics (2002) 30:233-237 What is the percentage of the different genotypes in the Finnish population? Enattah et al (2002) Nature Genetics (2002) 30:233-237 Conclusions: – What is the predominant phenotype in the Finnish population? – What does this tell you about evolutionary selection process at this locus? – (Why would this trait have been selected for in the Finnish population?) How could a SNP change the phenotype of an individual? – Develop some hypotheses • The SNP induces a change amino acid sequence? • The SNP causes a change in expression of the gene? • Anything else Possible avenues of exploration: • Look into how polymorphism effects gene regulation • Use data on reporter constructs? • What is an enhancer? Demographic data • What is the frequency of the SNP variant associated with lactose tolerance in the Finnish in other ethnic groups? – Use HapMap data as example • What is your prediction about lactose tolerance in these other groups? • What other information can students find on lactose tolerance demographics (use own families?) Populations being studied in Hapmap project: • 30 trios (two parents and an adult child) from Yoruba, Nigeria • 30 trios from Utah with European ancestry • 45 unrelated Japanese from Tokyo • 45 unrelated Chinese from Beijing http://snp.cshl.org/whatishapmap.html.en Allele Frequencies from NCBI and HapMap ALlele FREquency Database http://alfred.med.yale.edu/alfred/mvograph.asp?siteuid=SI00 1784U Examine Tiskoff data set from African populations – Does the same SNP determine lactose tolerance in these populations? – If not, what does this suggest about the evolution of the phenotype in these populations (idea of convergent evolution) – When did these variants arise? Example of Tiskoff data Subject Phenotype SNP 13910 genotype KEAA00 1 tolerant CC KEAA00 2 tolerant CC KEAA00 3 tolerant CC KEAA00 5 intolerant CC KEAA00 6 intolerant CC KEAA00 7 intolerant CC Floyd Reed and Sarah Tishkoff University of Maryland January 2007 LAB • Use PCR and RFLP to determine genotype of students at this C/T 13910 SNP – Students will isolate cheek cell DNA and use PCR to generate product containing SNP – Incubate PCR product with restriction enzyme Hinf1 – Run agarose gel to size fragments after digestion with Hinf1 – If T allele, PCR product will cut with Hinf and generate two fragments; product with C allele will not cut Create database of student results – Correlate ethnic background both genotype and phenotype – Data may be messy, but will accumulate over time • Extend discussion to importance of SNPs and individual predisposition to disease – See next slide for example deCODE Genetics • deCODE is a genetics company that has gathered genotypic and medical data from more than 100,000 volunteer participants in Iceland - over half of the adult population. They are using this information to find correlations between SNPs and diseases. • For example, scientists at deCODE Genetics and academic colleagues from the U.S. identified a SNP on chromosome 9 that confers increased risk of heart attacks. Of the 17,000 patients and control subjects in the study, more than 20% of participants carried two copies of the variant, which corresponded to an increased risk of more than 60%. • deCODE plans to use this type of information in the development of a DNA-based tests to identify individuals who are at elevated risk fro various diseases, thereby facilitating the implementation of preventive measures.