* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PPT
Genetic engineering wikipedia , lookup
Genomic library wikipedia , lookup
Ridge (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene therapy wikipedia , lookup
Genomic imprinting wikipedia , lookup
Public health genomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome (book) wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
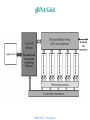
Parallel Detection of Regulatory Elements with gMP Bertil Schmidt, Lin Feng, Amey Laud, Yusdi Santoso Damayanti Gupta CMSC 838 Presentation Motivation Fundamental question How are expression levels of thousands of genes regulated ? Very important Understanding of gene function Response to environment Understand genetic causes of diseases Evaluate effects of drus Detect mutations Remember Sets of genes -> Pathways -> Genetic Networks Gene regulation Control decisions turn genes on/off Gene Regulation Network CMSC 838T – Presentation Talk Overview Overview of talk Motivation Technique Experiment Related work Conclusions CMSC 838T – Presentation Technique Motifs upstream of genes regulate gene expression Motifs are sites of regulatory activity Identify regulatory motifs by combining Gene expression data Detect common motifs occuring upstream of genes Huge datasets Utilise parallel computing CMSC 838T – Presentation Technique gRNA Java development framework gMP Java communication library REDUCE Algorithm to identify regulatory motifs REDUCE parallelised with gMP Increase computing power Get motifs ranked in statistical significance CMSC 838T – Presentation gRNA framework Consists of APIs CMSC 838T – Presentation gRNA - APIs Interact with data sources Provide functionality from biology Pipelines tasks into unified process Repository of resources Distributed programming CMSC 838T – Presentation gRNA environment gRNA Grid Clustered computing environment Application written for gRNA Multiple-tier application Applications operate from client computer Communicates with cluster through single computer Hosts EJB server Server identifies processing nodes each of these perform tasks CMSC 838T – Presentation gRNA Grid CMSC 838T – Presentation gMP Java based message passing tool Built on top of sockets Manages virtual processors to run on available machines Scalable Machines added/removed easily CMSC 838T – Presentation gMP Processes are grouped Communication primitives provided for sending and receiving data Collective communication to several nodes enabled modularly and efficiently Enables functions to be implemented on data CMSC 838T – Presentation REDUCE algorithm Based on model Upstream motifs contribute additively to expression level of each gene Quantify the extent to which these motifs contribute to expression data Fit log of expression ratio to sum of activating and inhibitory terms Find stastically most significant motifs Plots of fitting parameters suggest biological function CMSC 838T – Presentation REDUCE algorithm Terms Occurence vector Measure of how often a motif is found Expression vector Measure of gene expression CMSC 838T – Presentation REDUCE method Consists of 1) Motif frequency counter counts occurrences of DNA motifs upstream of each ORF motifs are about 7~11 nucleotides in length get occurence vectors CMSC 838T – Presentation REDUCE algorithm 2) Significant motif finder Use i) Normalised occurrence vector made for each motif nμ ii) Normalised vector of logs of gene expression ratio vectorsa Take dot product of these (a . nμ) ,and square. Can be considered as frequency of occurence X expressive power of regulatory motif It is squared to get rid of negatives Correlate gene expression with occurence of motif Largest dot product is most significant motif CMSC 838T – Presentation .... a is modified to remove effect of this motif residual gene expression vector Process repeated until motifs are ranked CMSC 838T – Presentation Table: Finding significant motifs Uses a - (.5816,.2522,.2886,-.5947, -.1595, -.3683) CMSC 838T – Presentation REDUCE parallelised with gMP... Parallel motif frequency counter Split set of ORFs equally Distribute across available nodes Each node calculates in parallel to get occurence vectors Matrix transposition Occurence vectors scattered across nodes Advantageous to store each vector in single node Transpose motif frequency matrix For each ORF can only calculate fraction of occurence frequencies for all motifs But the entire occurence frequency is needed CMSC 838T – Presentation ... Parallel significant motif finder Normalises occurence vector within each node At each node, most significant motif calculated Global most significant motif calculated Process iterated to rank occurence vectors Interface in gRNA allows ease of implementation CMSC 838T – Presentation Experiment Use Compaq Alpha system Consists of cluster of 8 AlphaServer SC/ES45 Connected by high-speed Alpha SC 16-Port switch and ELAN PCI adapter cards. Each server contains 4 Alpha EV68 processors CMSC 838T – Presentation Results Use 7090 gene expressions of yeast ORFs of length 600 Motifs upto length 7 Throughput (in MBytes/s) also shown 20 most significant motifs computed. CMSC 838T – Presentation Analysis Runtime scales well with number of processing nodes Frequency counter scales perfectly Motif finder also scales Cannot achieve perfect scaling because of communication overhead. CMSC 838T – Presentation Related work DiscoveryLink Provides configurable wrappers as interfaces to multiple data sources Kleisli system Systematically manages and integrates external databases Uses functional query language to perform correlation across databases Toolkits designed with functionality for specialised areas BioJava, BioPerl, PAL Sequence Analysis Ensembl initiative, DAS provide extensible approach to issue of annotating genomic data CMSC 838T – Presentation Related work Previous approaches using Java for high performance computing Bindings into native message-passing APIs(e.g.MPI) Does not allow easy integration into larger Java applications Pure Java message passing interfaces JMPI, CCJ Both implemented on top of Java RMI – Slower than using raw sockets CCJ tries to overcome – optimised RMI implementation – not portable Both cannot handle integration CMSC 838T – Presentation Comparison According to authors ... gRNA distinguishes itself Uses whole range of requirements for applications in computational biology Provides decoupled, yet inter-related subsystems Ease of 3rd party implementation CMSC 838T – Presentation Observations REDUCE surpasses traditional clustering approach REDUCE algorithm has high runtime Complexity depends on product of number possible motifs and that of genes. Grows exponentially with length of sequences So length of motif is restricted REDUCE algorithm is greedy suboptimal REDUCE is simplistic lacks parameters for interactions between motifs does not consider impact of other biological knowledge CMSC 838T – Presentation ... Not clear that results of REDUCE are biologically significant Experiment does not effectively show how higher computation power helps results Only analysis from 9 to 16 processors, is this sufficient to determine ‘good scaling’? CMSC 838T – Presentation Conclusions Finally... gRNA demonstrates efficient mechanism for development of genome-centric applications Further... Extensions to REDUCE have been proposed require higher computing power more specialised programming interfaces required Identifying communication patterns Use of data structures e.g. sequences, trees, matrices CMSC 838T – Presentation