* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Mining External Resources for Biomedical IE
Genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene desert wikipedia , lookup
Point mutation wikipedia , lookup
Gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene nomenclature wikipedia , lookup
History of genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Microevolution wikipedia , lookup
Helitron (biology) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
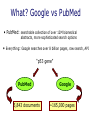
Mining External Resources for Biomedical IE Why, How, What Malvina Nissim [email protected] Why • goal: Named Entity Recognition • method: supervised learning • feature extraction • (text) internal features: word shape, n-grams, ... protein-indicative features: - of shape a0a0a0a… - followed by /bind/ - shorter than 5 characters • generalisations on training data might be incomplete • acquired evidence might be absent in test instance Getting Additional Evidence internal features might be insufficient, but good evidence might be somewhere else... • small and accurate lists of proteins (gazetteers) • use as rules • use as features • other texts might contain indicative n-grams • how to use other texts • which texts to use Note: some systems (MaxEnt for instance) can easily and successfully integrate a huge number of features How patterns “X gene/protein/DNA” “X sequence/motif” A. Create patterns (aim, method, input) B. Search corpus for patterns and obtain counts C. Use counts as appropriate Create Patterns (I) 1. AIM (granularity) distinguish entities from non-entities “X gene OR DNA OR protein” + bypass ambiguities and data sparseness – less information distinguish between entities “X gene” “X DNA” “X binds” + more information – ambiguities, data sparseness 1. AIM 2. METHOD 3. INPUT Create Patterns (II) 1. AIM 2. METHOD 3. INPUT 2. METHOD by hand (experts) + high precision, exact target – time consuming, experts needed automatically (collocations, clustering) + no human intervention – lower precision, not necessarily interesting patterns Create Patterns (III) 3. INPUT 1. AIM 2. METHOD 3. INPUT (“X gene”) low frequency words (as estimated from a non-specific corpus) words not found in standard dictionary NP chunks first output of classifier increase precision but lower recall all features – web prec .813 .807 rec .861 .864 f-score .836 .835 What? Google vs PubMed • PubMed: searchable collection of over 12M biomedical abstracts, more sophisticated search options • Everything: Google searches over 8 billion pages, raw search, API “p53 gene” PubMed 5,843 documents Google ~165,000 pages Google + PubMed “anything you want” site:<specific_site> “p53 gene” site:www.ncbi.nlm.nih.gov Rob Futrelle has this function available on this webpage: http://www.ccs.neu.edu/home/futrelle/bionlp/search.html • comment: sometimes PubMed reports “Quoted phrase not found” even when Google finds the phrase. PubMed provides phrase search only on pre-indexed phrases PubMed > Google • query expansion PubMed uses the MeSH headings to match synonyms (it will expand “Pol II” to search for “DNA Polymerase II”) Google will only try correct misspelling • field specific search PubMed allows field-specific searches (eg year) Google cannot refine its search in this respect • timeliness PubMed is updated daily Google is slow in updating PubMed > Google (cont’d) • ranking Google does a ‘vote’-based ranking: not necessarily good PubMed does not do any ranking (possibly bad too...) • truncation and flexibility PubMed accepts truncated entries and will look for all possible Variations. It will try break phrases if no matches are found. Google has a rigid search • manual indexing PubMed’s MeSH contain keywords not necessarily contained in the abstract Google cannot find something that is not mentioned in the abstract What What to to Use? Use? (or (or How How to to Use Use the the Evidence) Evidence) • as a rule + sure identification of entities – too powerful -> high risk of false positives might be better to use PubMed: less info but precise • as a feature + less false positives + some systems (MaxEnt) can integrate huge number of features – might still not get used or provide enough evidence might be OK to use Google: more info but not necessarily precise iHOP Nature Genetics, Vol. 36(7), July 2004 (Information Hyperlinked Over Proteins) A gene network for navigating the literature http://www.pdg.cnb.uam.es/UniPub/iHOP • uses genes and proteins as hyperlinks between sentences and abstracts http://www.pdg.cnb.uam.es/UniPub/iHOP • each step through the network produces information about one single gene and its interactions • information retrieved by connecting similar concepts • precision of gene name and synonym identification: 87-99% • readers can still check correctness of sentences when they are presented to them • shortest path between any 2 genes is on average 4 steps only