* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Promoter Analysis
Genome evolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
Molecular cloning wikipedia , lookup
List of types of proteins wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene expression profiling wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Community fingerprinting wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Molecular evolution wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene regulatory network wikipedia , lookup
Point mutation wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Transcription factor wikipedia , lookup
Gene expression wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Promoter (genetics) wikipedia , lookup
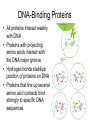
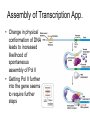
Promoter and Module Analysis Statistics for Systems Biology Transcription Factors • DNA binding proteins that facilitate or inhibit Pol II initiation or elongation • General transcription factors: – Used widely for many genes under many circumstances • Specific transcription factors – Used to initiate specific genes under specific circumstances • Distinction may not be so sharp! Transcription Factor Families • Several structures line up amino acids – Helix-turn-Helix (Homeodomain) – Helix-loop-helix – Zinc Finger • Mostly dimers • These families have proliferated because of their role in attracting transcription apparatus DNA-Binding Proteins • All proteins interact weakly with DNA • Proteins with projecting amino acids interact with the DNA major groove • Hydrogen bonds stabilize position of proteins on DNA • Proteins that line up several amino acid contacts bind strongly to specific DNA sequences Transcription Factor Recognition Sites • Typically 6-10 positions very selective and several others show bias • Often selectivity profile summarized by ‘motif’ Selectivity of Specific T.F.’s • Most TF’s recognize 6-10 bases of DNA • E. coli: longer (8-12 bp) TF’s – All sequences are effective • Yeast: areas around promoters selectively cleared of nucleosomes – ~ 30 x accessibility for those • Animal: cooperative binding of several T.F.’s Cofactors • Frequently the effect of DNA-binding proteins depends on co-factors • E.g. ER sits on the DNA but requires estrogen as a co-factor to function • Myc requires Max as a cofactor to stimulate transcription • If Max is coupled with Mad instead, the genes are repressed Assembly of Transcription App. • Change in physical conformation of DNA leads to increased likelihood of spontaneous assembly of Pol II • Getting Pol II further into the gene seems to require further steps The TF Family Circus Inferring Regulatory Architecture • Aim: to find which regulators influence gene expression • Concerns: – Contributions of many factors to any one gene • Approaches: – Decision tree (Computer Science) – Regression (more statistical) • DNA sequence motifs can be a surrogate The Israeli ‘Module’ Approach • Idea: model TF binding as a ‘decisiontree’ • Steps 1. Cluster gene expression profiles 2. Fit best regulator tree to each cluster 3. Re-assign genes to clusters • Iterate until converge Strengths and Weaknesses of Module Approach • Explicitly models interaction among regulators • Expression arrays give poor estimates of activity of TF’s or other regulators • Some regulators could repress genes • Discrete predictor model is inefficient Update: Estimating TF Activity • Since TF expression data is unreliable for activity, could we do better inferring TF activity? • Use DNA sequence motifs as surrogate for TF binding • Fit double E-M – complicated! The Regression Approach • Direct data on TF occupancy from ChIP • Two stages: – Find candidate TF’s by correlation between occupancy and sets of genes – Estimate TF activity in each condition by regression model Regression Steps Preliminary Screen r > rthreshold