* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene desert wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Protein moonlighting wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Point mutation wikipedia , lookup
Expanded genetic code wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Gene nomenclature wikipedia , lookup
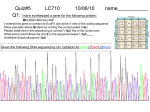
Quiz#3 LC710 9/29/10 name____________ Q1(1pt) Given the following protein: Nt- MNTSFSMLAAYMFLLIMLGFAINEAAFLTLYVTVLNYILLNLAVADEFRVFGGFTAT LYTSLGFFATLGGEIEALHWSLDVLAIERYVVVAIMGVAFTWVMALACAAVSLSFV IYMFVVHFIDIALRIVEIFFCYGMVIIMVIADFLICWELPYAGVAFYIFTHAFFAKKR TSAVYNPVIYIMMN-Ct Circle the potential transmembrane domains. There are more than 1. _______________________________________________ Q2(1pt) Given the following Kyte-Doolittle hydropathy plot: Circle on the number line where transmembrane regions reside. Q3(3pt) Given the following protein: MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIML * * = STOP You want to PCR up this conserved protein from an unknown species. -Design a fully degenerate oligo to the underlined Nt amino acids and include an extra EcoR1 site (GAATTC) at the 5’ end. This is a top strand oligo. -Design a fully degenerate oligo to the underlined Ct amino acids and include an extra BamHI site (GGATCC) at the 5’ end. This is a bottom strand oligo. Genetic Code: Write both oligos 5’ to 3’. Make sure to annotate 5’ and 3’ ends of your two oligos. (1pt) (2pt) Oligo 1:_______________________________ Oligo 2:_______________________________ _____________________________________________________ Q4(1pt) Given the following Gene below: Using the following list of symbols: ATG, STOP, SD, SA, 5’UTR, 3’UTR Completely label the gene below with these symbols KEY: SD: splice donor SA: splice acceptor 3’ UTR: untranslated 5’ ____________________________________________________ region (1pt) Extra Credit: AATAAA is a polyA recognition site. Using an arrow show approximate position in gene above.