* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Stanford Presentation, 10/23/2001
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
List of types of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Western blot wikipedia , lookup
Biochemistry wikipedia , lookup
Circular dichroism wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein domain wikipedia , lookup
Interactome wikipedia , lookup
Protein folding wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Homology modeling wikipedia , lookup
Protein adsorption wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
1
1
Sidechain Placement
and Protein Design
1
1
1
1
1
1
1
1
1
1
0
2
0
1
GCMB’07, 2 May
Protein design
• Sequence Structure Function
KDTIALVVST…
YPVDLKLVVKQ
Modify sequence
to change structure
and function [Looger03]
or behavior [Ambroggio06]
Ribose
binding
protein
TNT
binding
folding order
Protein Design or Redesign
• Create an amino acid sequence that folds to a stable protein
and performs a desired function
• Avoid:
– Sampling all sequences
– Solving protein folding
– Relying on molecular dynamics
• A successful design strategy:
build on an existing structure
– Scaffold: backbone from
a known folded structure
– Redesign ~20 residues
– Find side chains that fit
Outline
• Sidechain Rotamers & Rotamer Libraries
• Algorithms for Sidechain Placement
–
–
–
–
–
Brute Force
Dead End Elimination
Simulated Annealing
Stochastic Mean Field
Dynamic Programming
• A Biased View of Protein Structure & Design
– How is design done?
– Why is it successful?
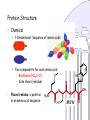
Protein Structure
• Chemical
– 1-Dimensional: Sequence of amino acids
– Two components for each amino acid
S
• Backbone (NCαC+O)
• Side chain (residue)
O
N
H2N
O
OH
• Placed residue: a position
in an amino acid sequence
HN
OH
OH
N
MSS
MSW
O
Side chain geometry
• Conformation flexibility from dihedral angles
– Side chain internal geometry
• Bond angles and bond lengths fixed
• Dihedrals c1, c2, … may rotate
• Rotamers: rotational isomers
N
N
– Side chains have preferred conformations
• Prefer dihedrals around 60o, 180o and -60o
• Rotamer Library: set of dihedral angles
[Ponder87, Dunbrack93, Lovel2000]
2
1
Side chain
conformation
side chains differ in size
(# of atoms)
and degrees of freedom
(# of c angles)
N
N
2
1
Serine c1 distribution
H
OH
H
Cα
Cα
Ni
Cβ
H
Ci
H
H
Ni
Cβ
OH
H
Ci
H
H
Cα
Ni
Cβ
OH
Ci
H
a chosen combination of side chain
torsion angles c1, c2, etc. for a residue
is known as a rotamer.
Side chain conformations--canonical staggered forms
Newman projections for c1 of glutamate:
O
O
C
g
b
C
d
CH2
CH2
CH
a
CO
CO
CO
c3
Hb
Cg
Cg
Hb
Hb
Hb
c2
N
Ha
N
Ha
N
Ha
c1
NH
Hb
Cg
c1 = 180°
c1 = +60°
c1 = -60°
t
g+
Hb
g–
O
glutamate
t=trans, g=gauche
name of conformation
Side chain angles are defined moving outward from the backbone, starting
with the N atom: so the c1 angle is N–Ca–Cb–Cg, the c2 angle is Ca–Cb–Cg –Cd ...
IUPAC nomenclature:
http://www.chem.qmw.ac.uk/iupac/misc/biop.html
Backbone independent rotamer library
• Dunbrack & Cohen, 1997
No. c1
No.
p
p|c1
c1
c2
What do rotamer libraries provide? [J. Meiler07]
• Rotamer libraries significantly reduce the number of
conformations that need to be evaluated during the search.
• This is done with almost no risk of missing the real
conformations.
• Even small libraries of about 100-150 rotamers cover about
96-97% of the conformations actually found in protein
structures.
• The probabilities of each rotamer in the library provide
estimates of the potential energy due to interactions within
the side chain and with the local backbone atoms, using the
Boltzmann distribution: E ln(P)
Side chain geometry
• Conformation flexibility from dihedral angles
– Side chain internal geometry
• Bond angles and bond lengths fixed
• Dihedrals c1, c2, … may rotate
• Rotamers : rotational isomers
– Side chains have preferred conformations
• Prefer dihedrals around 60o, 180o and -60o
N
N
– Rotamer Library: set of dihedral angles
[Ponder87, Dunbrack93, Lovel2000]
• http://dunbrack.fccc.edu/bbdep/bbdepdownload.php
(Backbone dependent and independent libraries)
• http://kinemage.biochem.duke.edu/databases/rotamer.html
(Backbone independent library)
2
1
Rotemers in crystallographic refinement
Fit structure to electron density from x-ray diffraction
Red indicate clashes w/
added hydrogen atoms
better choice
of side chain
& modified backbone
resolves clashes
Outline
• Sidechain Rotamers & Rotamer Libraries
• Algorithms for Sidechain Placement
–
–
–
–
–
Brute Force Search
Dead End Elimination
Simulated Annealing
Stochastic Mean Field
Dynamic Programming
Side Chain Placement Problem
• Given
–
–
–
–
–
–
A fixed protein backbone
A set of fixed (background) residues
A set of changing (molten) residues
A list of allowed amino acids for each molten residue
A rotamer library
A pairwise decomposable energy function
Find the assignment of rotamers to the molten
residues, S, that minimizes the energy function
Kinemage: rotamers for Ubiquitin surface residues
Energy Functions
• f: Protein Structure
– Lennard-Jones
• van der Waals attractive energies
• atom overlap repulsive overlap
– Electrostatics
– Solvent Effects
– Hydrogen bonds
a
b
__
__
- 6
12
d
d
c
__
d
• Often pairwise decomposable
– sum of atom-pair or rotamer-pair interaction energies
f=
f(i,j)
i<j
Side Chain Placement Problem
Find the assignment of rotamers to the molten
residues, S, that minimizes the energy function
min( Esingle(Si) + Epair(Si,Sj) )
SS
i
i<j
Functions stated in terms of rotamer energies
Esingle rotamer / background energy
Epair
rotamer pair energies
Side Chain Placement Problem
• NP-Complete
– Reduction from SAT [Pierce2002]
• Techniques
– Optimality Guarantee
• Dead-End Elimination [Desmet92, Goldstein94, Looger2001]
• Integer Linear Programming [Erickson2001]
• Branch and Bound [Gordon99, Canutescu2003]
• Dynamic Programming [Leaver-Fay2005]
– No Optimality Guarantee
• Genetic Algorithms [Jones94]
• Simulated Annealing [Holm92,Hellinga94,Kuhlman03]
• Self-Consistent Mean Field [Koehl96]
Dead End Elimination (DEE)
• Reduce the search space without losing the Global
Minimum Energy Conformation (GMEC).
• Eliminates rotamers which cannot be in the GMEC,
using more accurate (and more computationally
expensive) upper and lower bounds.
• Uses brute force search on rotamers remaining.
• Typically assumes that the scoring function can be
expressed as a sum of pair-wise interactions
A first, simple condition for elimination
• A rotamer can be eliminated for a residue when
the minimum (best) energy it obtains by
interaction with other rotamers
is still higher (worse) than the maximum energy
of some other rotamer:
score
ir
in
E ir min s E ir , j s min n [ E in max s E in , j s ] , i j
j
rotamer js interacted with
j
The Goldstein improvement
• A rotamer can be safely eliminated when there
exists a rotamer that has lower (better) energy
for each given environment.
• This criteria is more powerful, and typically
requires though more computational time.
score
is
it
E ir E it min s [ E ir , js E it , js ] 0 , i j
j
rotamer space
Even more powerful criteria can be obtained
with even more computation
• A rotamer can be safely eliminated when, for each
environment, there exists some rotamer that has
lower (better) energy.
score
rt
rs
rt’
rotamer space
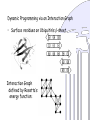
Dynamic Programming via an Interaction Graph
• Surface residues on Ubiquitin’s b-sheet
Interaction Graph
defined by Rosetta’s
energy function:
Interaction Graph
G = {V, E}, a multi-hypergraph
vertices molten residues
state space rotamers for a residue
edge possibility of residue interaction
scoring function interaction energy
Graph
Hypergraph
v
S(v)
e V
fe: S(v) →
ve
Interaction Graph Evaluation (Pairwise case)
• For G = {V, E}, min
Esingle(Si) + Epair(Si,Sj)
i
i<j
• Each vertex, v, has a function to capture interactions with
the background: f{v} : S(v) R
• Each pair of interacting vertices, {u, v}, defines an edge with
a function to capture pair interactions:
f{u,v} : S(u) x S(v) R
• Given an interaction graph, G={V,E},
find the state assignment S that minimizes SwVE fw
Bottom Up Dynamic Programming
Eliminate node v
• Let Ev be the edges incident upon v
• Let Nv be the neighbors of v
•
For each edge e Ev with scoring function fe,
let fe,v=s be edge e ’s scoring function with vertex
v fixed in state s
1. Create a new hyperedge incident upon Nv.
2. Compute fNv = min s S(v) e Ev fe,v=s
3. Remove v from graph
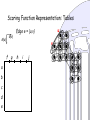
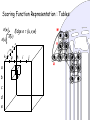
Scoring Function Representation: Tables
S(u)
S(v)
v
f g
a
b
c
d
e
u
Edge e = {u,v}
h
i
j
Scoring Function Representation : Tables
S(w)
S(v)
S(u)
l
kf
a
b
c
d
e
m
n
g
w
Edge e = {u,v,w}
v
o
h
i
j
u
Experiments and Results
• “Rotamer Relaxation Task”
– Sequence fixed – choose new rotamers for each residue
• “Redesign Task”
– Search of conformation and sequence spaces.
• Ubiquitin’s 15 surface residues
• Large rotamer library
– Relaxation, 32 states per vertex, tw-4 interaction graph
– Redesign, 680 states per vertex, tw-3 interaction graph
(drop one edge)
Running Time
Memory
Relaxation
~200 ms
(small)
Redesign
15.99 hrs
3.7 GB
Dynamic Programming for Hydrogen Placement
• Dynamic programming (DP) limited by treewidth of graph
instances
– Treewidths from graphs in protein design too large for DP to be
H
practical
• Adding hydrogen atoms to PDB
O
• Hydrogen placement via combinatorial optimization: REDUCE
[Word99]
– Non-pairwise decomposable energy function
– Previously used brute force
– Replaced with dynamic programming
• Interaction graphs have low treewidth
• Effective in practice: minutes to ms.
• REDUCE v3.02 in Molprobity suite, and distributed from
http://kinemage.biochem.duke.edu/software/reduce.php
Simulated Annealing
•
•
Stochastic optimization technique
–
–
–
Monte Carlo
Make a random change, determine ΔE
Metropolis criterion [Metropolis57]
• accept with probability
–
Gradually lower temperature T
In Side Chain Placement
_
___
ΔE
e
1
kT
ΔE > 0
o.w.
1. Assign each residue a rotamer
2. Repeat
1. Select a random residue, and a random alternate rotamer
2. Find ΔE induced by substituting the alternate rotamer
3. Accept/Reject substitution according to Metropolis criterion
Self-consistent mean field
• I planned to cull a description from Patrice’s
BioEbook sections:
– http://nook.cs.ucdavis.edu:8080/~koehl/BioEbook/design
_scmf.html
– http://nook.cs.ucdavis.edu:8080/~koehl/BioEbook/scmf.
html
but didn’t have time in class.
The practical problem of side chain modeling [M07]
• The way we deal today with the problem of protein
structure prediction is very different from the
way nature deals with it.
• Due to technical issues such as computation time
we are usually forced to accept a fixed backbone
and only then put the side chains on it.
• The quality of the side chain modeling is therefore
heavily dependent on the position of the backbone.
If the initial backbone conformation is wrong, the
side chain modeling quality will be accordingly bad.
• What is really needed is a “combined” algorithm
that optimizes backbone conformation
simultaneously with side chain modeling.
Protein Design or Redesign
• Create an amino acid sequence that folds to a stable protein
and performs a desired function
• Avoid:
– Sampling all sequences
– Solving protein folding
– Relying on molecular dynamics
• A successful design strategy:
build on an existing structure
– Scaffold: backbone from
a known folded structure
– Redesign ~20 residues
– Find side chains that fit
Why Design Proteins?
• Nature uses proteins
–
–
–
–
to
to
to
to
signal events
catalyze reactions
move cells (motors)
bear weight (I-beams)
• Design is an experiment to help understand folding/binding
• Industrial biosynthesis
– Proteins are both efficient and specific
• Cure disease
– Antibodies
– Inhibition peptides as drugs
• Perturb cell signaling pathways
Why do RosettaDesign, Dezymer, … work?
• Geometric approximations (3d jigsaw puzzles)
are surprisingly effective in design.
• They mine PDB structures for
behaviors of native proteins
and fragments.
• They precompute energies
for pairwise interactions.
• They use many fast computers
to allow detailed sampling
of discrete conformations.
• Fast optimization algorithms
• Competition
How do RosettaDesign, Dezymer, … fail?
• Computationally difficult to achieve good packing
and hydrogen bond satisfaction in protein core:
– Scores for packing, solvation and
hydrogen bond satisfaction
cannot be pairwise additive.
– Scores often used as filters;
we’d prefer to optimize.
• Stability of designed proteins
• Multistate or negative design
Protein Stability
• A naturally occurring protein adopts a compact
geometry when placed in water
• Stability is difference in free energies of the
folded and unfolded states
U
G
ΔG
F
Protein Stability
• A naturally occurring protein adopts a compact
geometry when placed in water
• Different proteins have different free energies in
their unfolded states
ΔG > ΔG
G
>
ΔG
Challenges in Protein Design
• Side chain placement is hard
– The complexities of individual instances of SCPP are
related to the treewidth of their interaction graphs.
• Tight, collision-free packing is often impossible on
the input scaffold
– The interaction graph to allow simultaneous optimization
of side chain and backbone structures
• Protein stability is not well captured by pairwise
decomposable energy functions
– The interaction graph supports using non-pairwise
decomposable energy functions during side chain
placement