* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download THE PUZZLING PROPERTIES OF THE PERMEASE (PPP) Kim …
Survey
Document related concepts
Rosetta@home wikipedia , lookup
Circular dichroism wikipedia , lookup
Structural alignment wikipedia , lookup
List of types of proteins wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein design wikipedia , lookup
Protein folding wikipedia , lookup
Protein purification wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Homology modeling wikipedia , lookup
Alpha helix wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Transcript
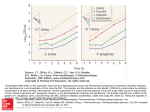
THE PUZZLING PROPERTIES OF THE PERMEASE (PPP) Kim Finer, Jennifer Galovich, Ruth Gyure, Dave Westenberg March 4, 2006 IS THERE AN E.COLI-TYPE IRON PERMEASE GENE (FepD) IN CHROMOHALOBACTER SP. AND IF SO, IS THE PERIPLASMIC-FACING RESIDUE SEQUENCE SIGNIFICANTLY DIFFERENT? BACKGROUND: Escherichia coli is known to have a ferric citrate transport system involving outer membrane permease, periplasmic transporters and an inner transmembrane protein encoded by the gene fepD. The fepD gene can also be found in other gram negative bacteria such as Pseudomonas aeruginosa PAO1. Chromohalobacter salexigens is a newly described species of bacterium able to survive in moderately saline environments. The species includes the previously described species, Halomonas elongata. One of its survival adaptations is the use of higher percentages of acidic amino acid residues in proteins that come into contact with the high salt environment. This modification protects the proteins from ‘salting out.’ HYPOTHESIS: We predict that transmembrane proteins such as FepD would also demonstrate this characteristic enrichment with acidic residues, but only in the hydrophilic regions facing the salty periplasm. Regions of protein hydrophilic regions facing the cytoplasm would be similar in charge makeup to the corresponding regions of FepD in E. coli, P. aeruginosa, etc. http://hugroup.cems.umn.edu/Research/Images_research/Ecoli.jpg These segments would be the same in the three species SALTY ENVIRONMENT, For Chromohalobacter These segments in Chromohalobacter would be more acidic than in the other two species METHODS We began by accessing an Excel spreadsheet of annotated C. salexigens sequences available at JGI. The spreadsheet was searched for ferric citrate transporter. The sequence reference was found (NP415122) and the homologous sequences from E. coli and P. aeruginosa were identified. The protein sequence for this accession was obtained from the JGI website. Protein sequences for FepD from E. coli K12 and P. aeruginosa (PAO1) were obtained from the NCBI website. Using Biology Workbench, we uploaded these three aa sequences and used GREASE to reveal the hydrophobicity profile (graph) for each protein chain. Using PI, we obtained the overall isoelectric point at each pH, with our interest being pH7 to compare the three proteins overall. Our hypothesis predicts that Chromohalobacter would have the lowest isoelectric point (more acidic residues). However, stronger evidence would be obtained by comparing each hydrophilic region in a pairwise fashion. TMAP allowed us to obtain a more precise prediction of which segments were hydrophobic vs. hydrophilic. There were 9 predicted transmembrane regions. Finally, each hydrophilic segment was submitted for PI analysis and the isoelectric point for each segment was compared among the three species. A third approach was to analyze the sequences using the Excel based analysis program, Protein Analysis. Protein analysis was used to plot the location of acidic amino acid residues along the entire peptide sequence. E. coli Data Sequence: MSGSVAVTRAIAVPGLLLLL IIATALSLLI GAKSLPASVV LEAFSGTCQS ADCTIVLDAR LPRTLAGLLA GGALGLAGAL MQTLTRNPLA DPGLLGVNAG ASFAIVLGAALFGYSSAQEQ LAMAFAGALV ASLIVAFTGS QGGGQLSPVR LTLAGVALAA VLEGLTSGIA LLNPDVYDQL RFWQAGSLDI RNLHTLKVVL IPVLIAGATA LLLSRALNSL SLGSDTATAL GSRVARTQLI GLLAITVLCG SATAIVGPIA FIGLMMPHMA RWLVGADHRW SLPVTLLATP ALLLFADIIG RVIVPGELRV SVVSAFIGAP VLIFLVRRKT RGGA Isoelectric Point: 10.476999 Kyte-Doolittle Hydropathy Profile Predicted Transmembrane Segments for E. coli: • • • • • • • • • TM TM TM TM TM TM TM TM TM 1: 2: 3: 4: 5: 6: 7: 8: 9: 11 - 39 (29) 66 - 86 (21) 94 - 114 (21) 121 - 143 (23) 152 - 173 (22) 193 - 221 (29) 243 - 271 (29) 283 - 303 (21) 310 - 330 (21) pI of Non-Transmembrane Segments Segment 1 2 3 4 5 6 7 8 9 Residues 1 – 10 40 – 65 87 – 93 115 – 120 144 – 151 174 – 192 222 – 242 272 – 282 304 - 334 pI 11.045 6.265 3.099 3.294 11.045 4.171 10.888 10.888 12.406 P. aeruginosa Data Sequence: MQASPMRRRR AVTLDALQAV VAGALMQALT SMGQYLGCAF AGLSVMLASL LLGWPGLAIG TWLLACLAVM QRWILPFSAL LGGPAFIVLV LRAWGLLAGA DPHDDRHLVV RNPLAEPGLL LGAGLAGIAV TGIIVLNAPP AGLAAAFALAA LLAGAATALAG IAAGLLLGAD RRFRLSRL Isoelectric Point: 11.822999 LLLALAALAS LALGSRPVPL RELRLPRTLV ALLAGAALG GINAGAALAV IVGVALFDLA FLLGQARETG TNPVRLVLAG EVFDRFRHWA AGSLSGSGFA RLNALALGQE I GQALGVDLRL PIAFVGLVAP HLARLLAGPD ILGRLLAAPT EIAAGIVALL Predicted Transmembrane Segments for P. aeruginosa: • • • • • • • • • TM TM TM TM TM TM TM TM TM 1: 11 - 39 2: 66 - 94 3: 100 - 120 4: 126 - 146 5: 156 - 184 6: 202 - 226 7: 246 - 274 8: 289 - 309 9: 316 - 336 (29) (29) (21) (21) (29) (25) (29) (21) (21) Kyte-Doolittle Hydropathy Profile pI of Non-Transmembrane Segments Segment 1 2 3 4 5 6 7 8 9 Residues 1-10 40-65 95-99 121-125 147-155 185-201 227-245 275-288 310-316 pI 11.04 5.24 3.29 5.92 6.01 11.04 4.07 11.04 3.29 C. salexigens Data Sequence: MLTRRTTRLA GLLAGLVLMA TTFAASVMLG ATLLHYDPSR VAHIIIVKER LPRAVIAVLV MQTLTRNPLA SPGILGINAG AMCFVVIAVA VWAALLGALV AACLVLMLSR GGGRAGPSSL AMFVSFSQGL LIIDHQSFES VLYWLAGSVS LPLFGIALLL CMLLVRHANA LMLGDDMVTS LLGLVVILLA GSSVALTGMI GFVGLIVPHM WLLPACALLG ACLLLLADVA SRFLMPPQDV TPFFIYLARR QQARP Isoelectric Point: 9.970999 TTELPPSTFI GASLAIAGTL LLPLHAPADY RVVLAGVAVT GRELSLVVPL LGMHAGTIKL ARGLFGFDHR PVGVMTALIG Predicted Transmembrane Segments for C. salexigens: • • • • • • • TM TM TM TM TM TM TM 1: 2: 3: 4: 5: 6: 7: 5 - 33 (29) 62 - 90 (29) 96 - 116 (21) 122 - 142 (21) 150 - 177 (28) 194 - 222 (29) 242 - 270 (29) • TM 8: 281 - 308 (28) Kyte-Doolittle Hydropathy Profile pI of Non-Transmembrane Segments Segment Residues pI 1 2 3 4 5 6 7 8 9 1–4 34 – 69 91 – 95 117 – 121 143 – 149 178 – 193 223 – 241 271 – 280 309 – 335 11.045 7.891 6.015 3.099 11.045 4.256 5.097 10.888 11.042 Comparison of pI Data Segment E. coli 1 11.045 2 6.265 3 3.099 4 3.294 5 11.045 6 4.171 7 10.888 8 10.888 9 12.406 P. C. aeruginosa salexigens 11.04 5.24 3.29 5.92 6.01 11.04 4.07 11.04 3.29 11.045 7.891 6.015 3.099 11.045 4.256 5.097 10.888 11.042 Protein Analysis Program plot of acidic amino acids - E. coli Charge / 28 Amio Acids Charge Along Protein Sequence 6 5 4 3 2 1 0 -1 -2 -3 -4 Total per 28 28 per. Mov. Avg. (Total per 28) 0 100 200 Amino Acid Residue Number 300 400 Protein Analysis Program plot of acidic amino acids - P. aeruginosa Charge Along Protein Sequence Charge / 28 Amio Acids 6 Total per 28 28 per. Mov. Avg. (Total per 28) 5 4 3 2 1 0 -1 -2 -3 0 100 200 Amino Acid Residue Number 300 400 Protein Analysis Program plot of acidic amino acids - C. salexigens Charge Along Protein Sequence Charge / 28 Amio Acids 4 Total per 28 28 per. Mov. Avg. (Total per 28) 3 2 1 0 -1 -2 0 100 200 Amino Acid Residue Number 300 400 CONCLUSIONS Hydropathy plots illustrate a similar topology for the FepD proteins of C. salexigens, E. coli and P. aeruginosa. We predicted that the loop domains or the C. salexigens FepD protein, exposed to the exterior of the cell would have a lower pI than the exterior domains of E. coli and P. aeruginosa. The calculated pI of the entire C. salexigens FepD protein is lower than the pI of the other two organisms. However, the calculated pI of individual loop domains and a plot of acidic residues show that the overall hypothesis is not supported. Several loop domains of the C. salexigens FepD protein have a lower calculated pI than the corresponding segments of E. coli, However, comparison to the corresponding domains of the P. aeruginosa FepP protein do not show the same trend. In fact, P. aeruginosa seems to have more acidic loop domains than C. salexigens.