* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Protein Architecture and Structure Alignment
Implicit solvation wikipedia , lookup
List of types of proteins wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein design wikipedia , lookup
Western blot wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein purification wikipedia , lookup
Protein folding wikipedia , lookup
Structural alignment wikipedia , lookup
Protein domain wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Alpha helix wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
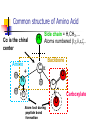
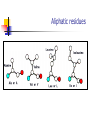
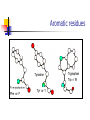
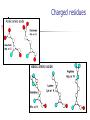
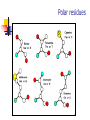
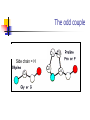
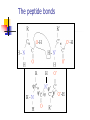
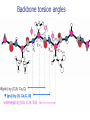
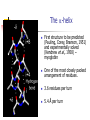
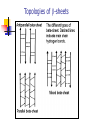
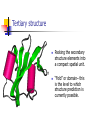
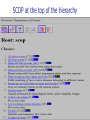
Part I : Introduction to Protein Structure A/P Shoba Ranganathan Kong Lesheng National University of Singapore Overview Why protein structure? The basics of protein Levels of protein structure Structural classification of protein structure Why protein structure? In the factory of living cells, proteins are the workers, performing a variety of biological tasks. Each protein has a particular 3-D structure that determines its function. ”Structure implies function”. Structure is more conserved than sequence. Protein structure is central for understanding protein functions. Sequence Structure Function To understand functions, we need structures α- conotoxin ImI and its three mutants Rogers et al., 2000, JMB 304, 911 Anfinsen’s thermodynamic hypothesis “The three-dimensional structure of a native protein in its normal physiological milieu (solvent, pH, ionic strength, presence of other components such as metal ions or prosthetic groups, temperature, etc.) is the one in which the Gibbs free energy of the whole system is lowest; that is, that the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment.” --- Anfinsen’s Nobel lecture, 1972 What drives protein folding? Hydrophobic effects Hydrophobic residues tend to be buried inside Hydrophilic residues tend to be exposed to solvent Hydrogen bonds help to stabilize the structure. Overview Why protein structure? The basics of protein Levels of protein structure Structural classification of protein structure The basics of protein Proteins have one or more polypeptide chains Building blocks: 20 amino acids. Length range from 10 to 1000 residues. Proteins fold into 3-D shape to perform biological functions. Common structure of Amino Acid Cα is the chiral center Amino H H + N R Side chain = H,CH3,… Atoms numbered b,g,d,e,z.. Ca H H Backbone O C - O Atom lost during peptide bond formation Carboxylate Aliphatic residues Aromatic residues Charged residues Polar residues S The odd couple Cg Cb Side chain = H Cd Ca Ca The peptide bonds Coplanar atoms Backbone torsion angles Ramachandran / phi-psi plot b-sheet a-helix (left handed) y a-helix (right handed) f Overview Why protein structure? The basics of protein Levels of protein structure Structural classification of protein structure Primary structure The amino acid sequences of polypeptides chains. Secondary structure Local organization of protein backbone: a-helix, b-strand (which assemble into b-sheet), turn and interconnecting loop. Ramachandran / phi-psi plot b-sheet a-helix (left handed) y a-helix (right handed) f The a-helix First structure to be predicted (Pauling, Corey, Branson, 1951) and experimentally solved (Kendrew et al., 1958) – myoglobin One of the most closely packed arrangement of residues. 3.6 residues per turn 5.4 Å per turn The b-sheet Backbone almost fully extended, loosely packed arrangement of residues. Topologies of b-sheets Tertiary structure Packing the secondary structure elements into a compact spatial unit. “Fold” or domain– this is the level to which structure prediction is currently possible. Quaternary structure Assembly of homo or heteromeric protein chains. Usually the functional unit of a protein, especially for enzymes Structure comparison facts Proteins adopt only a limited number of folds. Homologous sequences show very similar structures: variations are mainly in non-conserved regions. There are striking regularities in the way in which secondary structures are assembled (Levitt & Chothia, 1976). Overview Why protein structure? The basics of protein Levels of protein structure Structural classification of protein structure There are two major databases for protein structural classification: SCOP and CATH. They have different classification hierarchy and domain definitions. SCOP http://scop.mrc-lmb.cam.ac.uk/scop/ Structural Classification Of Proteins database Classification is done manually All nodes are annotated SCOP at the top of the hierarchy The hierarchy in SCOP Root Class Fold Superfamily Family Protein 5 classes: All-a, All-β, a/ β, a+ β, multi-domain Have the same major secondary structure & topological connections Probable common ancestry Clear evolutionary relationship CATH http://www.biochem.ucl.ac.uk/bsm/cath Class-Architecture-Topology-Homologous superfamily Manual classification at Architecture level but automated at Topology level The hierarchy in CATH 3 classes: Mainly-a, Mainly-β, a-β Class Architecture Overall shape as determined by orientations of secondary structures Topology Both the overall shape & connectivity of secondary structure Homologous Superfamily Sequence Share a common ancestor Classified based on sequence identity