* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Structural Insights into Kinase Inhibition Ramesh Sistla
Survey
Document related concepts
Catalytic triad wikipedia , lookup
Lipid signaling wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Biochemical cascade wikipedia , lookup
Signal transduction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Phosphorylation wikipedia , lookup
Drug discovery wikipedia , lookup
Ultrasensitivity wikipedia , lookup
MTOR inhibitors wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Citric acid cycle wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Transcript
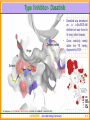
Structural Insights into Kinase Inhibition Ramesh Sistla and Subramanya H.S. Aurigene Discovery Technologies Ltd. #39-40, KIADB Industrial Area, Electronic City Phase II Bangalore 560 100 Kinases - Introduction • Kinases are enzymes catalyze phosphorylation • that ATP + protein = ADP + phosphoprotein • Key signaling enzyme • Human genome encodes > 500 kinases - Kinome • They have been implicated in different diseases including cancer, metabolic disorders and central nervous system indications. • Depending on the amino acid a kinase phosphorylates, they are known as Serine/Threonine or Tyorsine kinases. AURIGENE……Acccelerating Discovery www.cellsignal.com 2 Signaling Cascades • The figure shows the involvement of kinases in cell proliferation and survival. • In this cascade the phosphorylation of each kinase by its upstream kinase serves as a signal for downstream activity. • Inhibiting the pathway through inhibition of kinase involved in the pathway is an attractive proposition Current Medicinal Chemistry, 2008 Vol. 15, No. 29 3037 AURIGENE……Acccelerating Discovery 3 Promise of Kinase Inhibitors Some Advanced Kinase Inhibitors Druggable Genome Structure Identifier Target F F Kinome N N F O H N AMN-107 BCR/ABL N H N H 3C N N N N H N Imatinib CH 3 N O H 3C CH 3 N H STI-571 N BCR/ABL N Dasatinib CH 3 Cl O N H N S N N N H N N O O HC H N N BCR/ABL + OSI-774 Erbb OH CH 3 O BMS354825 CH 3 O CH 3 N • Kinases are an attractive target class – Druggability – Early successes (FDA approval of some of the kinase inhibitors) • Possibility of structure guided design – Large number of crystal structures in complex with inhibitors are available AURIGENE……Acccelerating Discovery 4 General Structure of Kinases N-terminal lobe • Bi-lobial structure • N-termial lobe – Mainly made of beta-sheets and connecting loops – One functionally important helix C-terminal lobe • Both lobes joined by a loop called hinge. • ATP binding pocket is in the interface between the lobes • C-terminal lobe – Mainly made of α-helices • Activation loop spans both Nand C-terminal lobes AURIGENE……Acccelerating Discovery 5 Important Structural Elements …GxGxxG… Helix-C Hinge • Plays an important role in catalysis Hinge – • Closes in on the ATP Helix C – • DFG……APE loop Glycine rich loop – • ATP Adenosine moiety of the ATP makes bidentate H-bond with this region Activation loop – Starts with conserved sequence DFG and ends with APE. AURIGENE……Acccelerating Discovery 6 Binding of ATP and Catalysis Orientation of the DFG motif critical for the phosphorylation H-bonds Hinge Metal Metal γ-phosphate coordinates with the metal S •Activation loop (DFG……APE) provides docking site for the substrate •Highly disordered and usually unresolved in the x-ray structures AURIGENE……Acccelerating Discovery T Substrate Y Phosphate 7 Important Residues Close up of the catalytic machinery N-terminal lobe Helix-C ATP Lys Salt bridge Asp Water Metal C-terminal lobe • • Glu In the active conformation of the kinases, a conserved Lys residue makes a salt bridge with a conserved Glu residue in the middle of the helix-C. This interaction ensures the positioning of the amino acid Asp (of the DFG motif) to coordinate with the γ-phosphate, the divalent metal ion and catalytic water molecule to facilitate catalysis AURIGENE……Acccelerating Discovery 8 Kinase Inhibitors • In most cases, inhibitors compete with ATP in order to inhibit the kinase – Such inhibitors are ATP mimetics in the sense that they make interactions similar to what ATP makes. G-loop Hinge ATP Inhibitor Ribose pocket Phosphate pocket ATP Inhibitor AURIGENE……Acccelerating Discovery 9 Various Subsites in Kinases Hinge Gatekeeper Deeper cavity Inhibitor Solvent ATP Schematic of the binding pockets PDB: 2C6E/1MQ4 An example of a kinase inhibitor bound in the ATP pocket is shown. Apart from hinge region interaction and solvent interaction, the inhibitor occupies a deeper hydrophobic cavity, also known as selectivity pocket Size of an amino acid preceding the hinge region controls the accessibility to the deeper pocket – Gatekeeper, (Typically Met/Leu/Thr/Ile/Tyr) AURIGENE……Acccelerating Discovery 10 Type I Inhibitor- Dasatinib • Deeper pocket Hinge • Dasatinib was developed as a c-Src/BCR-Abl inhibitor but was found to hit many other kinases. Cross reactivity mainly within the TK family; Approved by FDA Solvent 1nM 10nM 100nM 1μM 10μM Ref: Karaman et. al., NATURE BIOTECHNOLOGY VOLUME 26 NUMBER 1 JANUARY 2008 AURIGENE……Acccelerating Discovery 11 DFG-IN vs DFG-OUT Helix-C Gly rich loop DFG-Out DFG-In • The activation loop (DFG….APE) has to be IN when the kinase is active – DFG “in” conformation • The DFG loop has been shown to be in an “out” position when kinases are inactive. • This can be exploited in the design of inhibitors. AURIGENE……Acccelerating Discovery 12 DFG-IN vs DFG-OUT Helix-C ATP Gleevec DFG OUT DFG IN • Differences between DFG IN and DFG OUT structures are exemplified. • DFG loop in OUT position will clash with phosphate of ATP • When DFG moves to OUT helix-C also moves away creating the pocket shown by bold red arrow. • Gleevec binds to the DFG-OUT conformation of the C-Abl kinase. PDB:1T46 AURIGENE……Acccelerating Discovery 13 Example of Type-II Inhbition Hinge Phe-out conformation Schematic of the binding pockets PDB:1KV1 BIRB-796 binds to p-38 in the Phe-out conformation • The doublet of H-bonds with E-111 (helix-C) and D-207 (DFG loop) backbone is very important • Hence a urea or amide is the common feature in these inhibitors Ref: Karaman et. al., NATURE BIOTECHNOLOGY VOLUME 26 NUMBER 1 JANUARY 2008 AURIGENE……Acccelerating Discovery 14 R Some Known DFG OUT Inhibitors 2oo8 Tie – DFG out J.Med.Chem. 50: 611-626 2p4i Tie – DFG out Bioorg.Med.Chem.Lett. 17: 2886-2889 2osc Tie – DFG out R Bioorg.Med.Chem.Lett. 17: 2886-2889 N H 2og8 Lck – DFG out O 2ofv Lck – DFG out J.Med.Chem. 50: 611-626 2p2i KDR – DFG out Apart from a hinge binding group, the common feature in these molecules is existence of the bi-aryl amide/urea group which makes interaction with Glu (helix-C) and Asp (DFG loop) AURIGENE……Acccelerating Discovery 15 Allosteric Kinase Inhibition – Type III Helix-C ATP DFG loop • Certain kinases have an allosteric pocket in which an inhibitor can co-bind with ATP • The phosphorylation of the substrate is prevented by unavailability of the catalytic Asp • There are no hinge region interactions in these inhibitors. AURIGENE……Acccelerating Discovery 16 A Still Different Type of Inhibitor? • Recently Merck published the co-crystal structure of CHK1 kinase with an inhibitor that is bounds far away from the active site. • DFG loop is has IN conformation, but the inhibitor probably occupies substrate binding site. • Such inhibitors are not being designed yet. They could be results of HTS campaigns. PDB:3F9N AURIGENE……Acccelerating Discovery 17 SBDD at Aurigene All the structural biology efforts are to aid in more focused medicinal chemistry AURIGENE……Acccelerating Discovery 18