* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download ppt
Metalloprotein wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Proteolysis wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Lipid signaling wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Microbial metabolism wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Paracrine signalling wikipedia , lookup
Citric acid cycle wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Biochemical cascade wikipedia , lookup
Photosynthesis wikipedia , lookup
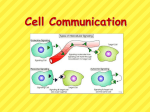
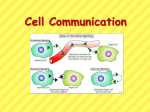
Glucose breakdown is exergonic! Energy is given off as heat and ATP. Redox reactions Occurs in matrix Net result: 4 carbon dioxide 2 ATP 6 NADH 2 FADH2 H pumped into inner-membrane space Oxygen is final electron acceptor Occurs on inner mitochondrial membrane – produces energy that drives oxidative phosphorylation Noncyclic electron flow Calvin cycle – end product is G3P (sugar) which will later be metabolized Into larger carbs Photorespiration: An Evolutionary Relic? • In photorespiration – O2 substitutes for CO2 in the active site of the enzyme rubisco – The photosynthetic rate is reduced – Energetically less efficient – Early atmosphere contained little oxygen • Enzyme rubisco could not distinguish between O2 or CO2 and thus today this continues • Or free radical preventer? C4 Plants • C4 plants minimize the cost of photorespiration (reduce uptake/use of O2) – By incorporating CO2 into four carbon compounds in mesophyll cells – Increasing CO2 levels • These four carbon compounds – Are exported to bundle sheath cells where O2 concentration is low, and then release CO2 used in the Calvin cycle. Mesophyll cell Mesophyll cell Photosynthetic cells of C4 plant leaf CO CO 2 2 • C4 leaf anatomy and the C4 pathway PEP carboxylase Bundlesheath cell PEP (3 C) ADP Oxaloacetate (4 C) Vein (vascular tissue) Malate (4 C) ATP C4 leaf anatomy BundleSheath cell Pyruate (3 C) CO2 Stoma CALVIN CYCLE Sugar Vascular tissue Figure 10.19 • CAM plants (cacti) – Open their stomata at night, incorporating CO2 into organic acids • During the day, the stomata close – And the CO2 is released from the organic acids for use in the Calvin cycle • Helps eliminate high water loss form stomata during heat of day (transpiration) • The CAM pathway is similar to the C4 pathway Pineapple Sugarcane C4 Mesophyll Cell Organic acid Bundlesheath cell (a) Spatial separation of steps. In C4 plants, carbon fixation and the Calvin cycle occur in different Figure 10.20 types of cells. CALVIN CYCLE Sugar CAM CO2 CO2 1 CO2 incorporated Organic acid into four-carbon organic acids (carbon fixation) 2 Organic acids release CO2 to Calvin cycle CALVIN CYCLE Sugar Night Day (b) Temporal separation of steps. In CAM plants, carbon fixation and the Calvin cycle occur in the same cells at different times. • Overview of cell signaling EXTRACELLULAR FLUID 1 Reception CYTOPLASM Plasma membrane 2 Transduction 3 Response Receptor Activation of cellular response Relay molecules in a signal transduction pathway Signal molecule Figure 11.5 • G-protein-linked receptors Signal-binding site Segment that interacts with G proteins G-protein-linked Receptor Plasma Membrane Activated Receptor Signal molecule Inctivate enzyme GDP CYTOPLASM G-protein (inactive) Enzyme GDP GTP Activated enzyme GTP GDP Pi Figure 11.7 Cellular response GTP replaced with GDP, now inactive • Receptor tyrosine kinases Signal-binding sites Signal molecule Signal molecule Helix in the Membrane Tyr Tyrosines Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Tyr Receptor tyrosine kinase proteins (inactive monomers) CYTOPLASM Tyr Dimer Activated relay proteins Figure 11.7 Tyr P Tyr P Tyr Tyr P Tyr P Tyr P Tyr Tyr P Tyr Tyr Tyr Tyr 6 ATP Activated tyrosinekinase regions (unphosphorylated dimer) 6 ADP Fully activated receptor tyrosine-kinase (phosphorylated dimer) P Tyr P Tyr P Tyr Tyr P Tyr P Tyr P Inactive relay proteins Cellular response 1 Cellular response 2 • Ion channel receptors Signal molecule (ligand) Gate closed Ions Ligand-gated ion channel receptor Plasma Membrane Gate open Think if these receptors as having allosteric activators/inhibitors Cellular response Gate close Figure 11.7