* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Access Slides
Cytoplasmic streaming wikipedia , lookup
P-type ATPase wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Circular dichroism wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
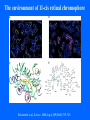
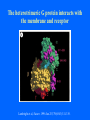
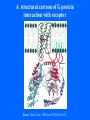
Structure of GPCRs and G proteins Goal of the lecture: Understanding the structural basis of how a GPCR activates a G protein Heterotrimeric G protein Pathway Clapham Nature. 1996 Jan 25;379(6563):297-299. Ribbon Diagram of Rhodopsin Structure Palczewski et al, Science. 2000 Aug 4;289(5480):739-745. Two dimensional Representation of Rhodopsin Palczewski et al, Science. 2000 Aug 4;289(5480):739-745. The environment of 11-cis retinal chromophore Palczewski et al, Science. 2000 Aug 4;289(5480):739-745. Salient features of Rhodopsin Structure Organization of the extracellular region serves as the basis seven-helix bundle arrangement 11-cis retinal holds transmembrane regions in the inactive conformation by interacting with key residues that participate in intra-helical interactions Activation of Rhodopsin Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Farrens et al, Science. 1996 Nov 1;274(5288):768-770. Design of the Experiment Mutate all Cys to Ser Bring back Cys of interest Construct double Cys mutants Keep Cys at 139 (helix 3) constant Vary 2nd Cys from 247-252 in helix 6 Put spin label on the Cys EPR spectroscopy Farrens et al, Science. 1996 Nov 1;274(5288):768-770. EPR spectra of inactive (dark state) shown as red trace and activated (meta-rhodopsin II) shown as yellow trace to study interactions between loops 3 and 6 Farrens et al, Science. 1996 Nov 1;274(5288):768-770. Results from EPR Spectroscopy Dark State: Distance between Cys at 139 and Cys at 248-251 = 12-14 Å After illumination increases in distances 23-25 Å Conclusion: Helices 3 and 6 move apart from each other after activation Biochemical Verification of EPR predicted movement of helices Cross link with disulfide reagent, cut with V8 protease Run SDS-PAGE If cross linked 1 band without DTT; 2 bands with DTT Farrens et al, Science. 1996 Nov 1;274(5288):768-770. Crosslinking of helices 3 and 6 blocks the ability of Rhodopsin to activate Transducin Fluorescence assay to measure GTPgS binding to transducin Farrens et al, Science. 1996 Nov 1;274(5288):768-770. Conclusions Helix 6 moves with respect to Helix 3 Movement is required for activation of transducin Helix 6 movement causes cytoplasmic loop3 to move Cytoplasmic loop3 is involved in coupling to transducin Farrens et al, Science. 1996 Nov 1;274(5288):768-770. G protein structure Lambright et al, Nature. 1996 Jan 25;379(6563):311-319. Space filling model of Ga interacts with Gbg Lambright et al, Nature. 1996 Jan 25;379(6563):311-319. The Gb interface that interacts with Ga contains key residues required for interaction with effectors Lambright et al, Nature. 1996 Jan 25;379(6563):311-319. G protein residues involved in regulation of effectors Space filling model of Gbg. Gb is white and Gg is pink. The green region is the area of Gb covered by Ga in the heterotrimer The smaller regions marked by colored dashed lines identify residues involved in interactions with various effectors. Each color corresponds to an effector Ford et al, Science. 1998 May 22;280(5367):1271-1274. In the heterotrimer the switch II region of Ga is contact with Gb Wall et al, Cell. 1995 Dec 15;83(6):1047-1058. Changes in the conformation of Ga in the GDP vs GTP bound forms and interactions with Gb Gb GTPgS-Ga Red GDP-Ga Blue Wall et al, Cell. 1995 Dec 15;83(6):1047-1058. The Switch II region of Ga has different conformation in the GDP and GTP bound states GTPgS GDP Wall et al, Cell. 1995 Dec 15;83(6):1047-1058. The heterotrimeric G protein interacts with the membrane and receptor Lambright et al, Nature. 1996 Jan 25;379(6563):311-319. A structural cartoon of G protein interaction with receptor Hamm J Biol Chem. 1998 Jan 9;273(2):669-672. Evolving view of receptors GPCRs exist as dimers Park et al, Biochemistry. 2004 Dec 21;43(50):15643-15656. Atomic Force Microscopy Picture of mouse rod-outer segment disc membrane Fotiadis et al, Nature. 2003 Jan 9;421(6919):127-128. Organization of the cytoplasmic surface of rhodopsin dimers are clearly visible Fotiadis et al, Nature. 2003 Jan 9;421(6919):127-128. Model of Rhodopsin Dimer Here phosphorylated Rhodopsin is shown binding to arrestin (This would be the Desensitized state) Park et al, Biochemistry. 2004 Dec 21;43(50):15643-15656. Model of rhodopsin dimer binding to one molecule of transducin Park et al, Biochemistry. 2004 Dec 21;43(50):15643-15656. Receptor Dimer Activation of G proteins Park et al, Biochemistry. 2004 Dec 21;43(50):15643-15656. A movie of this molecule is available from http://stke.sciencemag.org/cgi/content/full/sigtrans;2005/276/tr10/DC1