* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download A rule-based NLP pipeline for OWL
G protein–coupled receptor wikipedia , lookup
List of types of proteins wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
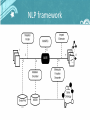
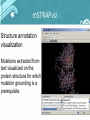
NLP pipeline for protein mutation knowledgebase construction Jonas B. Laurila, Nona Naderi, René Witte, Christopher J.O. Baker Background • Knowledge about mutations is crucial for many applications, e.g. Protein engineering and Biomedicine. • Protein mutations are described in scientific literature. • The amount of Information grow faster than manual database curation can handle. • Automatic reuse of mutation impact information from documents needed. Example excerpts "The W125F mutant showed only a slight reduction of activity (Vmax) and a larger increase of Km with 1,2-dibromoethane." • Mutation • Directionality of impact • Protein property "Haloalkane dehalogenase (DhlA) from Xanthobacter autotrophicus GJI0 hydrolyses terminally chlorinated and brominated n-alkanes to the corresponding alcohols." • Protein name • Gene name • Organism name Mutation impact ontology NLP framework Named entity recognition • Protein-, gene- and organism names – Gazetteer lists based on SwissProt – Mappings encoded in the MGDB • Mutation mentions – MutationFinder ~700 regular expressions – normalize into wNm-format Named entity recognition Protein Properties 1. Protein functions – Noun phrases extracted with MuNPEx – Activity, binding, affinity, specificity as head nouns 2. Kinetic variables – Jape rules to extract Km, kcat and Km/kcat in current implementation Mutation grounding Linking mutations positionally correct to target sequence • • 1. 2. 3. Important for reuse of mutation mentions Levels of grounding: mSTRAPviz Structure annotation visualization Mutations extracted from text visualized on the protein structure for which mutation grounding is a prerequisite. Protein function grounding • Mentions of protein functions are linked to correct Gene Ontology concepts. • Previously grounded proteins and mutations provide us with hints. • Grounding scored based on string similarity (later used during impact extraction) Relation detection • Impacts – Words describing directionality + protein properties • Mutants – Set of mutations giving rise to altered proteins • Mutant – Impacts – The causal relation between mutants and their impacts OwlExporter • Translates GATE Annotations to OWL instances • Application independent • Literature Specifications added automatically • Used here to populate our Mutation impact ontology to create a mutation knowledgebase Example query Retrieve mutations that do not have an impact on haloalkane dehalogenase activity (also retrieve the Swissprot identifier of the protein beeing mutated). Example query Retrieve mutations on Haloalkane Dehalogenase that do not impact negatively on the Michaelis Constant. Evaluation Mutation grounding performance What’s next? • Modularize into a set of web services • Database (re-)creation • Reuse in phenotype prediction algorithms, (SNAP)* *Bromberg and Rost, 2007 NLP pipeline for protein mutation knowledgebase construction Jonas B. Laurila Acknowledgement CSAS, UNB, Saint John [email protected] This research was funded in part by : Nona Naderi • New Brunswcik Innovation Foundation, New Brunswick, Canada CSE, Concordia University, Montréal [email protected] René Witte CSE, Concordia University, Montréal [email protected] Christopher J.O. Baker CSAS, UNB, Saint John [email protected] • NSERC, Discovery Grant, Canada • Quebec -New Brunswick University Co-operation in Advanced Education Research Program, Government of New Brunswick, Canada