* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download metabolomic and computational systems analysis
Genetic engineering wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Ridge (biology) wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Biochemical cascade wikipedia , lookup
Microbial metabolism wikipedia , lookup
Biochemistry wikipedia , lookup
Gene regulatory network wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Metabolomics wikipedia , lookup
Citric acid cycle wikipedia , lookup
Basal metabolic rate wikipedia , lookup
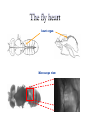
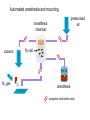
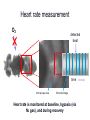
Metabolomic and computational systems analysis of hypoxic metabolism in Drosophila Jacob Feala1,2 Laurence Coquin, PhD2 Andrew McCulloch, PhD1 Giovanni Paternostro, PhD1,2 1) UCSD Bioengineering 2) Burnham Institute for Medical Research Cellular hypoxia response Hypoxia is the cause of cell death in many pathologies, mechanism not known All cells have intrinsic defenses Hypoxia tolerant organisms have highly orchestrated metabolic regulation Metabolic response is immediate and global Drosophila is hypoxia tolerant model Systems analysis of hypoxia response Complex balances must be maintained to tolerate hypoxia ATP supply and demand Redox potential Metabolic intermediates pH Hypothesis: flexible metabolic regulation key to hypoxia tolerance Systems biology to understand and model the complex control systems Hochachka, P. W. J Exp Biol 2003; 206:2001-2009 Drosophila as a model for hypoxia research Flies are hypoxia tolerant Simple system, genetic tools and libraries Genetic screen found gene required for tolerance 1 Hypoxia tolerance gene was successfully transferred to mammalian cells 2 human fly 1Haddad GG et. al., Proc Natl Acad Sci U S A. 1997 Sep 30;94(20):10809-12. 2Chen Q et. al., J Biol Chem. 2003 Dec 5;278(49):49113-8. Epub 2003 Sep 16. Phylogenetic tree General hypothesis for hypoxia tolerance Flexible metabolic regulation is the major source of hypoxia tolerance Immediate (minutes) Global (ATP production, biosynthesis, protein translation) Drosophila as a genetic model yeast Drosophila is the most genetically tractable model organism with a circulatory system. human No heart fly worm Phylogenetic tree • Humans and flies share many cardiac genes: – tinman (Nkx2.5) – ether-a-go-go (hERG) – troponin • Sequence similarity: Flies vs. humans 714 out of 929 (77%) of human disease genes had a Drosophila homolog (26 cardiovascular) (Reiter, 2001) Drosophila as a model organism Cardiac aging Cardiac hypoxia [From Giovanni’s slides] Fruit flies are highly tolerant to oxygen fluctuations Genetic screen for hypoxia tolerance: Adar (Haddad, 1997) Cardiac systems biology Several high-throughput datasets (protein interactions, genetic interactions, microarrays) Well annotated genome Ease of manipulation and short lifespan for phenotype screens Response to low O2 in flies and mammals Standard physiological endpoints for resistance to ischemia Infarct size: gold standard for mammals Myocardial stunning Recovery of mechanical function Post-ischemic arrhythmias Measurable in flies Sensitivity of fly heart to oxygen fluctuations not yet characterized Systems analysis of hypoxia response in Drosophila heart Regulation of metabolism is key in hypoxia-tolerant organisms Systems biology to understand and model the complex control systems Hypoxia Genes and Human Disease Apolipoprotein D Expressed in stroke, aging, alzheimers Defense against hypoxia-reperfusion injury in flies CD36 Fatty acid transporter Mouse KO had reduced ischemia tolerance (41% drop in cardiac output during ischemia) Our systems approach to modeling ATPgenerating metabolism: Metabolomics to find all anaerobic pathways Flux-balance analysis to simulate pathways under varying oxygen Generate novel, specific, testable hypotheses for hypoxia tolerance The fly heart heart organ Microscope view Automated measurement of heart function in Drosophila Gene Disruption Project Mail-order mutants (Bloomington, Indiana) Computer automated Detect and measure heart Anesthetize and mount on slide Breed and cross Automated cardiac phenotyping Automated anesthesia and mounting pressurized air anesthesia chamber fly vial vacuum N2 gas anesthesia slide computer-controlled valve Cardiac Mechanics Research Group Heart rate measurement O2 detected beat time microscope view M-mode image Heart rate is monitored at baseline, hypoxia (via N2 gas), and during recovery Preliminary Research: Cardiac phenotype Aim 1 Hypoxic cardiac phenotype Decrease in rate and fractional shortening, fast recovery Recovery is affected by %O2, duration Lots of data collected, needs better analysis Heart rate under 2% oxygen (one fly) 10 1/RR 120s hypoxia 5 0 0 20 40 60 80 time (s) 100 120 140 160 Environmental factors Measurement errors due to body movement 100 90 Percent completing measurement 80 70 60 Reflexive body contraction (loss of muscle tone?) below 2%, hard to measure heart Myocardial stunning at < 5% Increased response at high temperature 50 40 30 20 10 0 0 1 2 3 4 5 6 7 8 9 10 11 Percent oxygen Hypoxia and recovery normalized to baseline Cycle length in fly heart under 1 min of 3% O2 7 1.4 6 Baseline (20s) 1.2 Hypoxia (2 min) 5 Baseline (20s) Recovery (20s) 1 Hypoxia (1 min) Recovery (20s) 3 RR interval Normalized RR interval 4 2 0.8 0.6 1 0.4 0 0.2 -1 -2 1 2 3 4 5 6 7 Percent oxygen 8 9 10 11 0 24 26 28 30 Temperature (deg C) 32 34 36 1H NMR spectroscopy of hypoxic fly muscle • 0.5% O2 • 240 minutes • supervised by Laurence Coquin MAMMALIAN TISSUE: Troy H et. al. Metabolomics 2005; 1: 293-303 Global metabolic profile • Concentrations measured by targeted profiling (Chenomx): peak identification, alignment, subtraction • Lower confidence group due to spectra overlap Significant metabolites 1H NMR spectroscopy of flight muscle at t=0,1,10,60,240 minutes Reconstructing the Drosophila metabolic network Database integration KEGG: metabolic genes, enzymes, reactions, EC numbers, pathways Flybase: complete genome, proteins, function, compartment, mutant stocks, references Filtered gene index Pathways 109 EC numbers 437 Genes 1322 Genes (mitochondrial) 125 Genes (stocks available) 507 Reconstructing the network Network model of central metabolism 162 genes, 143 proteins and 158 reactions Includes glycolysis, TCA cycle, oxidative phosphorylation, β-oxidation, amino acids Elementally- and chargeStoichiometric balanced matrix Metabolic network reconstruction Literature and Databases Gene-protein-reaction associations Annotated Genome Reed JL et. al., Nat Rev Genet. 2006 Feb;7(2):130-41. Drosophila central metabolism Main energetic pathways in model Glucose NADH Acetate NH4 NADH α-Oxoglutarate Glycolysis ATP Glutamate ATP NADH Pyruvate Alanine Lactate Acetyl-CoA α-GPDH shuttle NADH Cytosol Mitochondria Pyruvate Acyl-carnitine shuttle FADH NADH CO2 Acetyl-CoA Oxaloacetate Known Drosophila pathways ATP Citrate TCA cycle NADH/FADH2 CO2 ATP O2 H2O Oxidative phosphorylation Hypothesized pathways Products seen in NMR NADH/FADH2 Flux-balance analysis Steady state assumption, flux constraints Optimize for objective function Mass and charge balance inherent Null Space of S ATP supply and demand Redox potential pH S matrix Metabolic network reconstruction Solution space Particular solution (optimal) Price, et. al. (2004) Nat Rev Microbiol 2, 886-897 Flux-balance analysis of hypoxia glc Simulation conditions ac lac ala - Glucose (and equivalents) only carbon substrate - Lactate, alanine, acetate constrained to NMR fluxes - Varied O2 uptake constraint - Objective: maximize ATP production Hypoxia simulation: key fluxes (Pseudo-) Mammalian Drosophila Stable pH Reduced glucose uptake Equivalent ATP Abbreviations: • atp: ATP production • co2: CO2 production • glc: glucose uptake • h: proton production • ac: acetate accumulation • lac: lactate accumulation • ala: alanine accumulation Flux-balance analysis of hypoxia glc Simulation conditions ac lac ala - Glucose (and equivalents) only carbon substrate - Lactate, alanine, acetate constrained to NMR fluxes - Varied O2 uptake constraint - Objective: maximize ATP production Cardiac phenotypes and single deletion analysis 16775 (red) 1.4 1.2 1 20862 (red) 1.4 0.8 1.2 0.6 1 0.4 0.8 0.2 0.6 0 0 20 40 60 80 100 0.4 0.2 0 0 20 40 60 80 100 WT vs mutant heart rate under hypoxia Colors represent effect of deletion on ATP production 11055 (red) 1.4 1.2 1 0.8 0.6 0.4 0.2 0 0 20 40 60 80 100 Conclusions Multiple anaerobic pyruvate pathways in fly may contribute to hypoxia tolerance New hypotheses to test: alanine and acetate production essential under hypoxia Systems modeling revealed emergent behavior Acknowledgements and many thanks Polly Huang Palsson lab, UCSD Bioengineering Adam Feist Thuy Vo Khoi Pham Questions