* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Histone depleted metaphase chromosomes Scaffold Attachment
Nucleic acid analogue wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Molecular cloning wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
X-inactivation wikipedia , lookup
Molecular evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
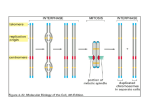
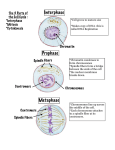
Centromeres Heterochromatin Kinetochore - spindle fiber attachment No universal DNA sequences Repeat sequences - CENs - 5 to 170 bp – humans, alphoid satellite repeats Specific associated proteins Telomeres Telomeres •About 15,000 bp in humans •Lagging strand problem •Telomere shortening •Hayflick – and then we die •Cancer, AIDs, stem cells The structure and organization of chromosomes change during the course of the cell cycle. Nonrandom chromosome positioning • Gene rich chromosomes toward center • Gene poor chromosomes toward periphery • Centromeres are not the determining factor • Chromosomes with adjacent positions more likely to interact cytolologically How porous are territories? • Previously it was predicted that active genes would be located at the surface of chromosome territories. • Now, it appears that they are also in the interior • Some are also found on loops outside of the territory In order for DNA to physically fit into a nucleus, several levels of packing are necessary. How is the DNA packaged in this way and how is it available for replication and transcription if it is tightly packed? The 4 nm DNA filament Histones • folding and coiling chromosomes • 45% of the total mass • 60 million molecules of each type per cell • Histones are highly conserved across all eukaryotic organisms • Histones are small basic proteins (102-135 aa) rich in lysine and arginine • Each histone contains a region that folds in a characteristic structure called the histone fold and a tail region • Tail region is post translationally modified in various ways to control many aspects of chromatin structure Core particle - Approximately 1.8 turns of DNA (consisting of 146 bp) wound around the outside of the histone octamer. Histone “tails” stick out between the coil of DNA Post-translational chemical modification of the tails controls function. Modification patterns comprise the “histone code”. The 10 nm nucleoprotein fibril Nucleosomes + 60 (approx) bp DNA A eukaryotic gene of 10,000 bp will be associated with about 50 nucleosomes. A human cell contains 3 x 107 nucleosomes. One molecule of histone H1 and the nucleososome is sometimes referred to as the chromatosome. Note that other chromatin modifying complexes include kinases, methylases and ubiquitin conjugating proteins. Acetylation typically correlates with transcriptional activation while deacetylation correlates with repression. • Non-histone proteins (NHPs, acidic proteins, nonhistone chromosomal proteins, NHC proteins) – help regulate DNA transcription and replication – at least 30 types “Chromatin remodeling complexes” and “Chromatin modifying complexes” are important for transcriptional activation Chromatin modifying complex Chromatin decondensation appears to require two types of protein complexes each made of several polypeptide subunits: 1. Histone acetylase complexes. These are often referred to as HATS for histone acetylases. 2. Chromatin remodeling factors. These are often refered to as Swi/Snf factors because they were first identified as yeast mutants defective in mating type switching and in the ability to metabolize sucrose (sucrose non-fermenting). Chromatin remodeling factors use energy from ATP hydrolysis to rearrange the packing of nucleosomes in higher order chromatin structures There are several different chromatin remodeling complexes in cells. Some of these bind to activation domains and de-condense the associated chromatin. Some bind to repression domains and condense the associated chromatin. Most interphase chromatin is condensed into 30nm coils. Most interphase chromatin is condensed into 30nm coils. A model for the structure of an interphase chromosome Lampbrush chromosomes (amphibian oocyte, immature eggs) In mitotic chromosomes, a scaffold is formed, and matrix attachment sites correspond to sites of chromatin attachment to the scaffold. Histone depleted metaphase chromosomes Scaffold Attachment Regions (SARs) Regions of the chromosomes with sequences specific for topoisomerase, HMG protein, and histone H1 binding Found only in untranscribed regions of the eukaryotic chromosomes Spaced along the chromosomes, with the intervening regions containing one or more genes? Highly AT rich (65%) and may be several hundred bp long Discrete loops of DNA appear to be connected at both ends to the scaffold Quaternary structure: final folding into chromosome shape