* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download A low resolution structure of a component of the Cytokine responsive
Spindle checkpoint wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Cytokinesis wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Phosphorylation wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
List of types of proteins wikipedia , lookup
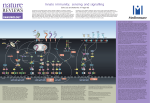
A Low Resolution Structure of a Component of the Cytokine Responsive Kinase Complex IKappaB Kinase (IKK) using SANS Joseph DiDonato Cleveland Clinic, Ohio, USA A Low Resolution Structure of a Component of the Cytokine Responsive Kinase Complex IKappaB Kinase (IKK) using SANS Thomas Tallant1, Xavier Lee1, Joe Zaccai2, Roland May2, Vitaliy Pipich3, Joseph DiDonato1,4* 1Department of Cell Biology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio 44195 USA 2Institute Laue-Langevin and EMBL Grenoble, France 38042 3Munich Outstation of Juelich Research Center, Germany 4Department of Cardiovascular Medicine, Cleveland Clinic, Cleveland, Ohio 44195 The IKappaB kinase (IKK) is a key regulator of the proinflammatory signaling cascade. It controls the release of the transcription factor NF-kB from its inhibitor molecule IkB via phosphorylation of conserved serine residues in IkB preferentially by the IKKbeta catalytic subunit triggering IkB’s polyubiquitination and degradation by the 26S proteasome. Once NFkB is freed from IkB, it translocates into the nucleus where it activates expression of its target genes acting as a key mediator of the proinflammatory gene response. IKK consist of two catalytic components, IKKalpha and IKKbeta, and a third structural component, IKKgamma. IKKgamma links upstream activation signals from the outer and inner cell membranes via signal-induced site-specific phosphorylation and/or K63-linked ubiquitination events on IKKgamma, leading to activation of the IKK complex. To date, there has been no experimentally-derived high or low-resolution structural information on the IKK complex or any of its components presented in the literature. Here we present for the first time structural information on the isolated recombinant IKKgamma subunit using SANS. The recombinant IKKgamma exists as a compact trimer which can be broken into a monomeric form using antibodies directed against its amino-terminus. Insight into the potential structure of IKKgamma in its “active” state was determined by SANS interrogation of two subdomains of IKKgamma that when expressed transiently together in trans in IKKgamma deficient cells restored basal IKK activation in the absence of stimulatory signals. These two subdomains interact strongly with each other in vitro as determined by surface plasmon resonance (SPR) using Biacore. Each subdomain individually forms a homo-dimer with itself and when mixed with each other preferentially forms a hetero-dimer. SANS-determined shape of the heterodimeric IKKgamma subdomains reveals an open extended structure compared to the compact structure of the IKKgamma subunits present either in the IKK trimer or the monomeric IKKgamma subunit in the monomeric IKKgamma-antibody complex. This closed to open conformation of IKKgamma may indicate how posttranslational modifications of IKKgamma in response to cell signals may trigger structural conformational changes in IKKgamma that lead to IKK complex activation of the catalytic subunits IKKbeta and IKKalpha.