* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download SF 106 year 1 report 2010
Gene expression programming wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
History of genetic engineering wikipedia , lookup
Essential gene wikipedia , lookup
Public health genomics wikipedia , lookup
Genome evolution wikipedia , lookup
Pathogenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Ridge (biology) wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene expression profiling wikipedia , lookup
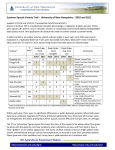
Project Title: Developing breeding and selection tools to reduce spoilage of soft fruit and wastage in the supply chain Project Number: SF 106 (HL 0195) Project Leader: Julie Graham Scottish Crop Research Institute Mylnefield Invergowrie Dundee DD2 5DA Tel: 01382 562731 Project partners: Fax: 01382 562426 SCRI, Delta-T, HDC, Marks & Spencer, Mylnefield Research Services, ReDeva, Thomas Thomson (Blairgowrie) Ltd Report: First annual report, 2010 Previous reports: None Key Personnel: Dr Mary Woodhead, Dr Craig Simpson, Dr Paul Hallett, Dr Heather Ross, Dr Blair McKenzie, Dr Christine Hackett, Dr Linda Milne, Dr Julie Graham Project co-ordinator: Peter Thomson (Thomas Thomson Ltd) Date Project commenced: July 2009 Expected completion date: June 2012 Key words: soft fruit, wastage, spoilage, softening, fruit quality, shelf-life, molecular markers, breeding, selection tools Whilst reports issued under the auspices of the HDC are prepared from the best available information, neither the authors nor the HDC can accept any responsibility for the inaccuracy or liability for loss, damage or injury from the application of any concept or procedure discussed. The contents of this publication are strictly private to HDC members. No part of this publication may be copied or reproduced in any form or by any means without prior written permission of the Horticultural Development Company. 1 AUTHENTICATION We declare that this work was done under our supervision according to the procedures described herein and that the report represents a true and accurate record of the results obtained. [Name] [Position] [Organisation] Signature ............................................................ Date ............................................ [Name] [Position] [Organisation] Signature ............................................................ Date ............................................ Report authorised by: [Name] [Position] [Organisation] Signature ............................................................ Date ............................................ [Name] [Position] [Organisation] Signature ............................................................ 2 Date ............................................ CONTENTS Page No Grower Summary Headline 4 Background and expected deliverables 4 Summary of the project and main conclusions 4 Financial benefits 5 Action points for growers 5 Science section Introduction 6 Results and Discussion 7 Conclusions 12 Publications and technology transfer outputs 12 Exploitation plans 12 3 Grower Summary Headline A good start has been made to the process of identifying genes responsible for fruit softening in raspberry. Background and expected deliverables Demand for UK fruit currently outstrips supply. Fruit softening remains the main cause of post harvest waste and lost revenue in all soft fruit. Fruit firmness is essential to maintain quality, enabling fruit to withstand extended storage before being transported across the UK. This is in addition to the 7 days of shelf-life demanded by supermarkets. A 1-2 day improvement in fruit shelf-life would increase the value of harvested fruit and reduce waste. This project aims to identify genes associated with fruit softening, by using the Glen Moy x Latham mapping population, which is already an established and successful resource for QTL mapping. The Glen Moy x Latham raspberry map, the gene resources developed in current LINK project HL0170 and the latest sequencing technologies (454 Life Sciences, Roche), will be used in this work. Having identified genes responsible for fruit softening, their response to abiotic stresses (eg.temperature and water) will be investigated. The most robust genes/markers can then be exploited in future breeding programmes to breed varieties with reduced fruit softening characteristics. Summary of the project and main conclusions Work in the first year of the project has used the genetic map of the Glen Moy x Latham cross to develop screening protocols to identify genes which confer fruit firmness and good shelf life. Work has also been undertaken to improve our understanding of the stresses that occur in the supply chain and how they relate to fruit firmness scores. 4 Using a number of genes already known to be linked with fruit softening, work in the first year has been undertaken to improve our understanding of the forms in which they occur. Financial benefits This work will confer significant financial benefits to the industry: The volume of fruit as food waste going to landfill in the UK will be significantly reduced. Financial savings in the retail industry alone could reach £2.5 million annually for soft fruit, with additional savings at the farm. Increased shelf-life will further enhance the reputation of UK fruit as a high quality product, leading to improved fruit sales. Action points for growers There are no action points at this stage. 5 Science Section Introduction Demand for UK fruit currently outstrips supply. Fruit softening remains the main cause of post harvest waste and lost revenue in all soft fruit. Fruit firmness is essential to maintain quality, enabling fruit to withstand such extensions to storage time before transport across the UK. This is in addition to the 7 days of shelf-life demanded by supermarkets. A 1-2 day improvement in fruit shelf-life would increase the value of harvested fruit and reduce waste. This project aims to identify genes associated with fruit softening, by using the Glen Moy x Latham mapping population, which is already an established and successful resource for QTL mapping. The Glen Moy x Latham raspberry map, the gene resources developed in current LINK project HL0170 and the latest sequencing technologies (454 Life Sciences, Roche), will be used in this work. Having identified genes responsible for fruit softening, their response to abiotic stresses (eg.temperature and water) will be investigated. The most robust genes/markers can then be exploited in future breeding programmes to breed varieties with reduced fruit softening characteristics. The project objectives are: Objective 1. Identify map locations for softening phenotypes (QTLs) from phenotypic analyses that mimic stresses in the supply chain. Objective 2. Carry out large scale sequencing of genes expressed in ripening raspberry fruit. Objective 3. Identify sequence polymorphisms in 8 named genes and other fruit-related genes such as MADS box genes (Obj. 2) implicated in fruit ripening/softness. Objective 4. Generate an updated linkage map with fruit-related genes and identify genes that co-locate with softening QTLs. 6 Objective 5. Study gene expression profiles and potential alternative splicing events in key softening genes (Obj. 4). Objective 6. Validate robust genetic markers and gene sequences (Obj. 5) by comparison with raspberry germplasm (and with other members of the Rosaceae and grape). Results and discussion Good progress has been made against milestones during the first year: 1.1 Develop screening protocols for fruit firmness and shelf life and assess material from the Glen Moy x Latham cross. Completed Sept 2009. 1.2 Understand stresses in the supply chain and how they relate to firmness scores. Ongoing-to be completed Aug 2011. 2.1 Extract RNA from raspberry white/red and red fruit of both Glen Moy and Latham for 454 sequencing. Completed August 2009. 3.1 Isolate and identify sequence polymorphisms in 8 named genes and other fruit-related genes (Obj. 2) implicated in fruit softness. Completed April 2010. 1.1 Develop screening protocols for fruit firmness and shelf life and assess material from the Glen Moy x Latham cross. Fruit softness was evaluated using two procedures. Firstly, fruit was assessed by a subjective (‘breeder’) score of firmness on the bush on a 1 - 4 (Firm – Soft) scale for the 188 progeny from the Glen Moy x Latham cross replicated in triplicate at an open field site and polytunnel site at SCRI. Secondly, ripe fruit (15 berries) were collected in plastic punnets from 1 and 2 replicates from each of the field and polytunnel sites, respectively. The weight of 10 berries per progeny was measured and the softness was determined on 6-8 individual berries using various measurements with a QTS-25 Texture Analyzer (Figure 1). 7 Figure 1. The QTS-25 Texture Analyzer. Statistical tests were used to determine correlations between the breeder score of fruit firmness and all the quantitative measurements from the machine. Preliminary analysis revealed a significant correlation between the breeders score and the machines calculations of Final load, Hardness, and Hardness1 work done. There were significant differences (genetic variation) among the Glen Moy x Latham mapping progeny for the traits, Final load (p = 0.005), Hardness (p = 0.005), and Force/Mass_N_kg (p < 0.001). The Mass (weight of 10 fruit) value also varied significantly between the progeny (p < 0.001). Mapping analysis was used to collect all trait data and assign to a Rubus genetic linkage map. Preliminary analysis showed that calculations for Final load, Hardness, and Force/Mass_N_kg, together with the breeders score were significantly more heritable and all showed QTLs on LG3. The most significant markers on LG3 associated with these QTLs were P14M61-121 (AFLP marker) and Rub120a (Rubus microsatellite DNA). Thus, both the breeders score and some of the Texture Analyzer data can be used to the identify soft and firm phenotypes within the Glen Moy x Latham mapping population and can be used to identify chromosomal regions responsible for trait variation. Table 1. Data collected from QTS-25 Texture Analyzer 8 Note: definitions for each selected calculation are available. 1.2 Understand stresses in the supply chain and how they relate to firmness scores. Information about stresses in the supply chain from the grower (T. Thomson Blairgowrie Ltd.), marketing organization (ReDeva) and multiple retailers (M&S) will be obtained and data from fruit assessments will be compared to predictions from the developed protocol. Work to be initiated although several discussions have taken place and the use of calibrated shaking incubators for holding punnets, and to follow or mimic the ‘transport’ of fruit from grower to supermarket by using sensors for assessment of compression and impact damage are options to be explored. This objective will be further discussed at the next consortium meeting. 2.1 Extract RNA from raspberry white/red and red fruit of both Glen Moy and Latham for 454 sequencing. Total RNA was extracted from white/red (pre-ripe) and red (ripe) fruit of both parents Glen Moy and Latham and complementary DNA (cDNA) was synthesised. The cDNA was sent to the 9 University of York for 454 sequencing. The 454 data will be used to create an extensive sequence database of genes expressed in raspberry fruit (‘fruit transcriptome’), and in the first instance, the database has been searched for candidate genes related to fruit softening to be used in objective 3.1. A repeat run of 454 sequencing for both Latham white/red and red fruit samples was necessary and the procedure has now generated over 1.3 million sequences across the four samples (Table 2). The size distributions of the sequences obtained were similar for each sample (400-500 bases). It has been a long process to assemble the many different sequences of this Rubus dataset into meaningful and related groups to facilitate further analysis. Table 2. The numbers of mRNA reads and total length of sequence obtained from each 454 run after quality trimming during sequence assembly. Parent Sample* White/Red Moy Red Moy White/Red Latham 1 Red Latham 1 White/Red Latham 2 Red Latham 2 Total Number of mRNA Reads 788562 136595 23821 7653 189938 179795 1326364 Total Length (MBases) 225.9 41.2 7.2 2.3 55.8 53.9 386.3 *A repeat run was carried out for both Latham samples, and the first run and second runs are designated by 1 and 2, respectively. 3.1 Isolate and identify sequence polymorphisms in 8 named genes and other fruit-related genes (Obj. 2) implicated in fruit softness. Candidate genes with expected roles in the fruit softening process were identified using inhouse (Rubus) and public sequence databases together with 454 datasets, and were used to identify sequence polymorphisms (insertions/deletions [indels] or single nucleotide polymorphisms [SNP]) in Glen Moy and Latham. The procedure involved the PCR amplification of candidate genes, sequencing of the products, and sequence alignments to identify any polymorphisms between the two parents and in 6-12 selected progeny to confirm the polymorphism segregated and therefore could be mapped. 10 Sanger sequencing or Pyrosequencing procedures were used to identify the SNPs, whereas indels were identified using a standard genotyping protocol and an ABI 3730 capillary sequencer. Data for a number of genes were mapped onto the SCRI Rubus genetic linkage map using the JoinMap programme and their locations are shown in Table 3. Although the final analysis for several candidate genes has yet to be completed, interestingly, βgalactosidase1 (β-gal1) and expansin1 (Exp1) mapped onto LG 3, linked to the location of QTLs for fruit softening determined from the breeders score of fruit firmness and some Texture Analyzer measurements. Table 3. Status of mapping of candidate genes implicated in cell wall disassembly and fruit firmness. Candidate gene Identifier Glen Moy x Latham linkage map group (LG) Pectinmethylesterases (PME) ERubLR_SQ9.2_C12 LG 1* Polygalacturonase (PG) ERubLR_SQ13.4_H10 LG 5* Cellulase2 (Cel2) RiCellulase LG 5* β-xylosidase ERubLR_SQ01F_D13 LG 7* Xyloglucan ERubLR_SQ5_3_H01 LG 7* Expansin4 (Exp4) RiExpansin LG 7* Pectate lyase (PL) RiPL LG 7 β-galactosidase1 (β-gal1) CL1386Contig1 (454) LG 3 β-galactosidase2 (β-gal2) CL2657Contig1 (454) Pyrosequencing β-galactosidase3 (β-gal3) CL5915Contig1 (454) Pyrosequencing Expansin1 (Exp1) ERubLR_SQ13.2_E09 LG 3 Expansin2 (Exp2) CL6813Contig1 Genotyping Arabinofuranosidase (Arab) CL6475Contig1 (454) Pyrosequencing Cellulase1 (Cel1) CL5428Contig1 (454) LG 1 Pectinmethylesterases CL3991Contig1 (454) Genotyping endotransglycosylases (XET) (PME) Polygalacturonase (PG) ras3_PG_Moy_Contig 29 & Primers designed 11 60 (454) *Mapped in collaboration with other projects. ERubLR – indicates sequences held in the database derived from Latham root cDNA library from HortLink HL0170. No changes to the project have been required to date. Conclusions None can be drawn at this early stage of the project. Publications and technology transfer outputs None to date. Exploitation plans The aim of this work is to develop robust assisted breeding and selection tools that will enable breeders to accelerate new variety development with extended shelf-life and thus reduce fruit spoilage and waste in the supply chain. No exploitation plans can be drawn up at this early stage of the project. 12