* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Micro 071023
Metabolic network modelling wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Biosynthesis wikipedia , lookup
Butyric acid wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Photosynthesis wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Biochemistry wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Citric acid cycle wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Electron transport chain wikipedia , lookup
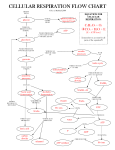
Microbes as Energy Transducers The Metabolic Menu Metabolic Strategies Respiration & Fermentation Chemolithotrophy Photoautotrophy Biogeochemical Cycles Metabolism in Primitive Organisms The tendency of a molecule (B) to accept an electron from another molecule is given by its electrode potential (E), a.k.a the reduction potential. E0 is the electrode potential under standard conditions. Eh is the actual electrode potential. E0’ is the electrode potential at pH 7 (under biological conditions). Molecules with more negative Eh or E0’ values are better electron donors. Electrons flow spontaneously to molecules with more positive Eh or E0’ values. The total free energy change occurring in these two half reactions can be calculated by the following equation: DGo’ = (-nF)(DEh) where Eh is the free energy change under standard biological conditions, n is the number of electrons transferred, and F is the faraday constant (-96.48 kJ/V) DEh = Eo’(oxidized partner) - Eo’(reduced partner) Oxidation and Reduction are Coupled Reactions Negative E0’ A becomes oxidized, losing electrons Positive E0’ B becomes reduced, gaining electrons Redox Rxns: Eo’= -0.42 V Eo’= +0.82 V DGo’ for the formation of water: Eo’of electron donor once oxidized DEh = (+0.82) – (-0.42) Eo’of electron donor when reduced DGo’ = (-2)(96.48)(1.24) = -239.27 kJ Other final electron acceptor O2 as final electron acceptor Metabolic Menu For Chemotrophs a.k.a chemolithotrophs, chemoautotrophs a.k.a chemoorganotrophs, chemoheterotrophs Electrons in redox reactions are carried from the primary electron donor to the terminal electron donor. Electrons are not freely diffusible in the cell. They are carried by electron carriers. Electron carriers 1. Freely diffusible (coenzymes/cofactors) – move electrons from one enzyme to another to take part in reactions (like batteries or electron taxis) -NAD/NADH: catabolic, energygenerating reactions, transfers hydride ion (2 e- + 1 H+) -NADP/NADPH: anabolic, biosynthetic reactions 2. Fixed carriers, where carrier is a prosthetic group firmly attached to enzyme in the cytoplasmic membrane (electron transport chain)… like a power line or stops on the city bus line Then, need to store energy… Energy currencies for cells are usually compounds whose DG0’ is greater than 30 kJ/mol. Anhydride bonds Energy contained in glucose used to convert low-energy phosphoryl groups to highenergy bonds: 1,3 bisphosphoglycerate Phosphoenolpyruvate Acetyl-CoA Succinyl-CoA 2 stages: 1. Splitting glucose (C6) into two phosphoglyceraldehydes (C3) 2. Oxidizing phosphoglyceraldehyde to pyruvate Two Ways to Make ATP: Quick & Dirty Fermentation or Turbo-Charged Aerobic and Anaerobic Respiration Fermentation: Balances the reduction of NAD+ with a subsequent oxidation of NADH. Restores electrical balance. Net yield: 2ATP and “waste” products that are derived from the original electron donor. ATP Synthase Structure & Function F1 subunits: 3 a, 3 b, 1 g, 1 d, and 1 e. b subunits = site of ATP hydrolysis/synthesis F0 subunits: proton channel Torque generated rotor stator F1 Subunit Topview ATP Synthase acts as a rotary motor turning in 120 degree steps. 3 or 4 protons flowing through turns c proteins enough to change conformation of b protein 1 ATP Conformation of active sites on b subunits is constantly changing… like watching The Wave at a Mariners game. Loose Open: substrate binding Tight: product release from highaffinity site, actually requires more energy than ATP synthesis The force that powers ATP Bidirectional: synthase is the proton motive force, or Dp. Major route of H+ into cell, but protons can get pumped out, too. ATP synthase reaction is close to equilibrium; direction depends on Dp One site for ATP hydrolysis and synthesis (Protons can also be pushed back out by reverse e- flow, usually in chemolithotrophs R- + K+ K+ R- K+ K+ K+ + + - K - K K+ K+ K+ R K+ K+ + K valinomycin + K+ + K K K+ K+ K+ K+ R R- Uncouplers are ionophores that collapse the Dp. They inhibit ATP synthesis that is coupled to e- transport. Other cations could run ATP synthase too… Judicious choice of ionophores (small lipidsoluble, cation-carrying, ion gradient-thwarting molecules) show that you can run ATPase with a K+ gradient. Valinomycin, added to cells loaded with K+, uncouples Dp by carrying out K+ and creating a temporary diffusion potential. Net gain …in the rich world of the gut where E. coli lives! Bacteria e- source Archaea e- sink A theory on a uniting feature of the Archaea: energetically stressed lifestyle. Archaea are adapted to energy-poor extremes, but Bacteria outcompete them in “easy” environment. Success in Stress: Marine sedimentary record of large radiation of tetraether-based, isoprene membranes in an anoxic period during the mid-Cretacean, perhaps driven by competition for ammonia (e- donor) utilization? Doesn’t prefer life of luxury: Low extant diversity (89 genera vs. 1,400 bacterial genera) due to inability to radiate in replete environments Energetic stress: Halophilic lifestyle (must prevent excess Na+ from entering cell) Thermophilic lifestyle (must keep from melting/denaturing) Nitrifiers (low N, low O2 in open ocean) Anaerobic methanogens (always Archaea) (not a good living: one methane produced can gain as little as one cation (1/3 to ¼ ATP) Aerobic methanotrophs Acidophilic lifestyle Alkaliphilic lifestyle – think what this does to one’s Dp!! -Na+ powered flagellae and ATP synthase -local H+ gradients (proton pump next to ATPase? -S-layers, membrane properties retain H+ Archaea are good at maintaining cells. Bacteria prefer the “growth” lifestyle. Few or no Archaeal pathogens because living in a host is not adapted to a lowenergy ecosystem Archaea survive heat, low pH; Bacteria prefer cool and neutral D.L. Valentine, 2007, Nature Reviews Microbiology 5: 316 Membranes: Stable to temperature, pH Less permeable to ions so less leakage/uncoupling; better maintenance of chemiosmotic potential D.L. Valentine, 2007, Nature Reviews Microbiology 5: 316 Speaking of stress… Fermentation, a very stressful situation O2 is not highly soluble in water Many environments easily become anoxic Decomposition of organic material must continue anaerobically If alternate e- acceptors (SO4-2, NO3-, Fe3+) are not available, then… Fermentation! ~3 ATP per glucose now, not 38 as in aerobic respiration. Respiration vs. Fermentation ADP + Pi ½ O2 NADH + H+ B NADH + H+ FADH2 FAD H2O NAD+ ATP NAD+ BH2O Fermentation – Key Features (1) Always anaerobic (if O2 around, can’t be used) (2) No externally supplied terminal electron acceptor. (3) Substrate-level phosphorylation is the rule*. Many types.... 2 major themes (1) NADH+H+ gets oxidized to NAD+ (2) Electron acceptor is usually Pyruvate or its derivative. *Rules are always meant to be broken! Pasteur Effect: ~20X more biomass when aerated Redox balance The short list Fermentation as a diagnostic tool: end products are a metabolic fingerprint Escherichia, Salmonella, Proteus, Aeromonas: Ferment glucose to a number of organic acids. Produce H2 and CO2: enzyme called formic hydrogenlyase splits formic acid to make these. Enterobacter makes neutral fermentation products. Fermentation as a diagnostic tool: end products are a metabolic fingerprint Enteric bacteria: g-proteobacteria, G- rods, nonsporulating, facultative anaerobes, oxidase-negative, fermentative Enterobacter: negative Escherichia, Salmonella, Proteus, Aeromonas: positive Methyl Red Test: Red if pH < 5.0 Escherichia, Salmonella, Proteus, Aeromonas: negative Voges-Proskauer Test: pink = butanediol produced Enterobacter: positive Clostridium spp: endospore-forming, anaerobic rods Ferment amino acids, purines, aromatic compounds/antibiotics Pyruvate Tricarboxylic acid (TCA) cycle: used in aerobic respiration, anaerobic respiration, and even fermentation! Don’t want NADH or FADH2 Still need precursors for amino acids etc. Convert from oxidative pathway to reductive pathway (incomplete CAC) Blocked between a-ketoglutarate and succinyl-CoA, cannot operate in oxidative direction Runs in reverse from OAA to Succinyl CoA Fumarate dehydrogenase instead of succinate dehydrogenase Reverse arrows for NADH and FADH2 Propionic Acid Fermentation: incomplete citric acid cycle Pyruvate OAA Lactate Mal Variations in many fermentative microbes Fum Succ Succ CoA Prop CoA MethylMalonyl CoA The unusual fermentation of succinate Sodium ATPase: Uses Na+ gradient …DG0’ so small (-20.5 kJ) that cannot even do substrate-level phosphorylation! Coupled to sodium transport; slowly build up “equity” in form of Na+ gradient (Dp) The unusual fermentation of oxalate H+ ATPase: Uses proton gradient …DG0’ so small (-26.7 kJ) that cannot even do substrate-level phosphorylation! Oxalate consumes protons in cytoplasm during conversion to formate. Slowly boosts Dp by making cytoplasm more basic.