* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download פרויקט מחקר - בנימין קפא
Vectors in gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Primary transcript wikipedia , lookup
Holliday junction wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Protein moonlighting wikipedia , lookup
Frameshift mutation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Non-coding RNA wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Microevolution wikipedia , lookup
Genomic library wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Homologous recombination wikipedia , lookup
Point mutation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
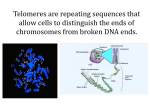
The genetic interactions between telomerase and CST proteins in Kluyveomyces lactis Binyamin Kaffe The Telomeres The end regions of eukaryotic chromosomes. • These regions are composed of a repetitive sequence of double stranded DNA, ending with a single stranded 3’overhang. • Insure that no gene will get lost during replication when the chromosomes shorten. • Protect the end of the chromosome from deterioration or from fusion with neighboring chromosomes. • Telomeres usually elongate by telomerase. However In some cases they elongate via an alternative pathway (ALT) which involves recombination. The Telomerase • A ribonucleoprotein (RNP) containing an RNA molecule (TER), which serves as a template, and the telomerase reverse transcriptase protein (TERT), which synthesizes the DNA upon the RNA template. • Active in unicellular organisms, mammalian germ cells and most cancer cells. Not active in most human somatic cells. Human template Model organism Kluyveromyces lactis A budding yeast 6 chromosomes – 12 telomeres Has a long and exact telomeric repeat of 25 repetitive bases - CGGATTTGATTAGGTATGTGGTGT The telomerase RNP & accessory proteins in budding yeast • Protein subunit TERT – Est2 in • Double strand binding proteins – Rap1, • • budding yeast RNA subunit TER – Tlc1 in Saccharomyces cerevisiae Rif & Sir family Single strand binding proteins –the CST complex (Cdc13-Stn1-Ten1) and more…. Important domains in K. lactis TER Template- a complementary sequence for the telomeres Pseudoknot- stabilizes the template The different stems are important for the functionality and binding of different proteins (ex. TWJ binds the TERT subunit) CST complex • Recruits telomerase by its • • • interaction with the telomerase protein Est1. Coordinates the synthesis of the telomere by its interaction with DNA polymerase. Protects it from degradation by exonuclease action. Functions as a negatively regulator of the telomere length. General Objective To understand the genetic and functional interactions between Cdc13 & Stn1, and the RNA subunit of telomerase. Specific objectives To check the telomere phenotype and viability of K. lactis strains overexpressing CST proteins in the background of: • TER1 (encoding the K. lactis TER) deletion. • TER1 mutated in Reg2, an element possibly involved in the recruitment of the telomerase to the telomere. • TER1 mutated in the pseudoknot, which is known to affect the activity of telomerase, but it is not assumed to affect the recruitment of telomerase. Plasmid insertion by electroporation Methods Serial streaks Morphology observation Southern blot The plasmids TER containing plasmids • CEN-ARS plasmids with WT, mutated or no TER • gene. All of the introduced TERs have a single mutation in the template region. (In order to detect the newly synthesized telomere with a specific probe, and to set a newly recognition site for the restriction enzyme BclI which enables to evaluate the difference between the old part of the telomere and the new one.) • All the newly introduced plasmids have a HIS1 selectable marker. The yeast strain contained a WT TER (URA3) plasmid that was negatively selected by seeding on 5-FOA plates. Over expression (o.ex.) plasmids •2µ plasmids with a high copy number. • All the newly introduced plasmids have a URA3 selective gene. The mutations used in the project Previous lab observation WT 1 telomere 2 telomeres 7 telomeres Reg2-L2 The telomeres do not show any BclI incorporation [no BclI digestion and no telomeres were detected when hybridized with BclI probe (not shown)] which means that the new template was not used. However telomere length is similar to WT telomere length, although the chromosomes replicated ~120 times. The genomic DNA was digested with EcoRI or EcorI+ BclI and analyzed by Southern blotting and hybridization a WT telomere radioactive probe. Questions How could it be that the new telomerase template was not used but the telomeres maintained their WT length? •Is the ALT pathway active? The telomeric bands do not show a long smear after the hybridization with a telomeric probe which is typical for the ALT pathway Maybe the over expression of Cdc13 or Stn1 regulates the length of the telomeres to WT? •Alternatively, is there another mechanism that maintains the telomere original length even so the telomerase is not active? 1st attempt results Colony morphology observation Phenotype scores of colonies from 6 passages. Scores are between 1-4, where 1 indicates a round and smooth colony and 4 indicates the most aberrant looking colonies. Data in the table are weighted averages of numerous colonies in 2 clones. Empty Vector Stn1 o.ex. Cdc13 o.ex Streak #1 #2 #3 #4 #5 #6 #1 #2 #3 #4 #5 #6 #1 #2 #3 #4 #5 #6 WT Ter 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 terD 4 4 3 2.5 1.5 1.5 4 1.5 2.5 3 3 3.5 Reg2-L1 2 1 1.5 2 1.5 1.5 2 2.5 1.5 2.5 2 1.5 1.5 1 2 2 1.5 1.5 Reg2-L2 1.5 1.5 3 1.5 2 1 2.5 2 3 2 2 2 2 2 2.5 2 2.5 1.5 N.A. contamination Notable is the improvement of the morphology during the passages which is due to the elongation of the telomeres in the alternative pathway. In the case of terD the improvement started after the 1st passage, in the Reg2 mutants which have a non-optimal telomerase the phenotype worsened first due to the telomere shortening and the improvement started in later stages. In some strains we see the phenotype worsening again after improving, since the recombination is suppressed when telomeres are long, and activated when they shorten. There is no distinct difference when the proteins are overexpressed. telomeric length in Reg2 ter mutations is shorter then WT, however the overexpression of the CST proteins has no significant effect on the length. The smear seen in terD/EV is typical for recombinant pathway survivors due to the lack of tight regulation of telomeric length. No telomeric bands were detected in the lanes of terD/o.ex Cdc13, it appeared to be a contamination. Southern blot (The BclI incorporation enables to evaluate the difference between the old part of the telomere and the new one by BclI digestion and specific hybridization) Conclusion These results disconfirm the previous lab results; the overexpression of the CST proteins has no significant effect on the length of the telomeres. However… The previous lab results were obtained in a somewhat different procedure. The plasmid shuffling by plating on a 5FOA plate in order to negatively select the cells that lost the WT plasmid (URA3), was done prior to any of the transformation and not after the first transformation. Could the lack of any active telomerase for ~20-30 generations induce an alternative pathway and lead the cells overexpressing CST proteins to a different pathway? We decided to redo the experiment with this difference of removing the WT TER prior to transforming a mutant ter. 1st attempt WT TER1 (URA3), 2nd attempt Different TER1s (with BclI site) (HIS1), O.EX. plasmid 2nd attempt results The WT TER1 was removed by plating on 5FOA prior to transforming a mutant ter. • This attempt did not include the L1 Reg2 mutation which leads to significant similar results as the L2 mutation as we saw in the 1st attempt. • The pseudoknot (PK) mutation was included in order to check if the CST proteins have an effect in a case were the catalytic activity of telomerase is abolished but there is no putative effect on the recruitment of the telomeric proteins. Colony morphology observation ‘A’ clones Streak Streak WT /EV 0 1 2 3 4 5 6 WT /EV WTWT /Stn1 /Stn1 WT WT/Cdc13 /Cdc13 ⌂ ⌂ /EV /EV ⌂ ⌂ / Stn1 / Stn1 ⌂/ ⌂/ Cdc13 Cdc13 • The WT strains morphology is round throughout all the streaks. • In the case of a severe mutation (terD) the morphology is severe in the early passages and it improved during the passages which is believed to be due to an elongation of the telomeres in a recombination pathway. The telomeres shorten again after a few passages as we see that in some strains (e.g. passage 6 of D/ Stn1) the phenotype worsens after the improvement, which is due to the fact that the recombination is suppressed when telomeres are long, and activated when they shorten. ‘A’ clones Streak Streak 0 1 2 3 4 5 6 Reg2 Reg2 /EV /EV Reg2 Reg2 /Stn1 /Stn1 Reg2 Reg2 /Cdc13 /Cdc13 A PK PK /EV /EV PK PK /Stn1 /Stn1 PK PK /Cdc13 /Cdc13 • In the reg2 mutation which is not as severe, the phenotype worsened first due to the telomere shortening and the improvement started in later stages. From these photos it seams like the overexpression of Cdc13 prevents the colonies from fully returning to a round phenotype (additional photos in next slide). • In PK mutation which is a severe mutation we see similar behavior to the ter deletion strain, however it does seam like Cdc13 prevents the colonies from fully returning to a round phenotype . Colony morphology of Reg2/Cdc13 Reg 2 /Cdc 13 A Reg 2 /Cdc 13 B Reg 2 /Cdc 13 C Notice a difference between the different clones, which may indicate variability in the survival pathway: •In ‘A’ clones the colonies basically maintained the same phenotype throughout the passages (passage 3 is an exception were we observe two phenotypes). •In ‘B’ clone we see worsening and improvement in passage 5, and again worsening in passage 6. •In ‘C’ clones we could observe a large diversity in colony morphology and two cycles of worsening and improving. ‘A’ clones No evidence to BclI sites incorporation in PK mutants. Some of the strains show smears indicating the recombination pathway. Southern blot The different smear patterns seen in terD is typical for recombinant pathway survivors due to the lack of tight telomeric length control, and the fact that the telomeres shorten again after the recombinant elongation. In L2 mutants we observe BclI incorporation and what seems like regulated smears (Smears maybe more regulated since there is a use of the new template). Stn1 shows longer telomeres than EV and Cdc13 which maybe due to the regulation. Cdc13 shows the same length as EV, however there is an extra fragment at 4kb which could be due to excessive recombination. Not possible for a terD to have BclI incorporatio n maybe misplaced by another strain. ‘B’ clones Smear patterns are different than in clones A. The recombination undergoes regulation, and therefor the telomeres shorten again after the recombinant elongation. Patterns are different at different stages No BclI site incorporation in Reg2/Cdc13! However, we do not observe a telomere length which is similar to the WT length, as observed previously in the lab Conclusions • In case where the WT TER1 gene (Ura3) was negatively selected only after the • • transformation of the mutated ter (His1), there was no effect of overexpressing the CST proteins. In case were the WT TER1 gene (Ura3) was negatively selected prior to the transformation of the mutated ter we did observe some variance between the L2 strains and in one case we observed no use of the mutated template when Cdc13 was overexpressed. However we did not observe the phenotype that was observed previously in the lab. There are short smears in the L2 strains, which we believe to be generated by recombination. Since the elongation in this pathway is induced only when the telomeres shorten, we have cycles of telomeres shortening-elongating by recombination-shortening again…. During this cycle the length could be a WT length as we see in some strains in my results, and that may explain the previous lab result. The reason that in one case there was no use of the mutated template is not clear, but may indicate competition between telomerase and the recombination pathway. I.e., recombination was induced by the overexpression of Cdc13 and suppressed telomerase activity. Future plans • To check the different reg2/Cdc13 clones that show different growth behaviors. • Check the phenotype in different passages in order to observe the effect through the generations.