* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download File - interACT - American College of Toxicology
Therapeutic gene modulation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Essential gene wikipedia , lookup
Metagenomics wikipedia , lookup
Microevolution wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome (book) wikipedia , lookup
Epitranscriptome wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Pharmacogenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Minimal genome wikipedia , lookup
Genomic imprinting wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Ridge (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Public health genomics wikipedia , lookup
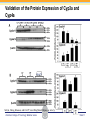
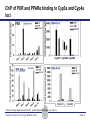
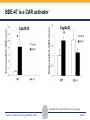
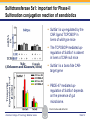
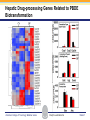
Novel Interactions between Gut Microbiota and Host Hepatic Xenobiotic Biotransformation – Lessons Learned from the Germ-free Mice Julia Yue Cui, PhD Assistant Professor, Sheldon Murphy Endowed Chair in Toxicology and Environmental Health Department of Environmental and Occupational Health Sciences, University of Washington Three examples of the role of gut microbiome in regulating the hepatic drug-processing genes • Gut microbiome and liver development • Probiotics/conventionalization on hepatic drugprocessing gene expression • Interactions between gut microbiome and environmental chemicals American College of Toxicology Webinar series Slide 2 Liver P450 expression modulated by gut microbiota Selwyn, Cheng, Bammler, Prasad, Vrana, Klaassen, and Cui JY*, 2015 Toxicol Sci, 147: 84-103 American College of Toxicology Webinar series Slide 3 Protein expression was altered in livers of Germ-free Mice as compared to Age-matched Conventional Mice Selwyn, Cheng, Bammler, Prasad, Vrana, Klaassen, and Cui JY*, 2015 Toxicol Sci, 147: 84-103 American College of Toxicology Webinar series Slide 4 Four Developmental Patterns of Critical Drug Processing Genes in Livers of Conventional and GF mice A Selwyn, Cheng, Bammler, Prasad, Vrana, Klaassen, and Cui JY*, 2015 Toxicol Sci, 147: 84-103 American College of Toxicology Webinar series Slide 5 Age and gut microbiota affect hepatic drugprocessing genes • The absence of gut microbiota produced age-specific effects on the regulation of hepatic drug-processing genes • The age-specific changes may be due to different types/ratios of intestinal bacteria at various ages during development in CV mice American College of Toxicology Webinar series Slide 6 Regulation of Hepatic Drug-metabolizing Enzymes in Germ-free mice by Conventionalization and Probiotics • Probiotics: live microorganisms that confer a health benefit to the host when administered in adequate amount • VSL3 is a combinatorial probiotic that is used for human intestinal disorders, such as inflammatory bowel disease and ulcerative colitis American College of Toxicology Webinar series Slide 7 Introduction of exogenous bacteria to germ-free mice: Expression of hepatic drug-processing genes • Age-matched 2-month-old adult conventional (CV) and germ-free mice (males, n=5~8 per group) were administered 4.5×106 CFU/ml drinking water for 28-days • In a separate experiment, 1-month old GF mice were taken out of the isolator and housed with the feces from CV mice for 2-months • All mice were 3-month-old prior tissue collection. American College of Toxicology Webinar series Slide 8 a" a" 0.20 0.15 0.10 b" 0.05 b" 0.00 C l 3 rGF 3 GGF+ F ol L CV+ LGF+ trCV to ex CV on V VSVSL3 on F VSVSL3 C C G C GF V 1.0 Average Cq=19.02 Cyp3a44 a" 0.8 0.6 b" b" 0.4 0.2 c" 0.0 c" l 3 ro 3 GGF+ F ol LCV+ CV GF LGF+ ntr VSVSL3 nt VSVSL3 ex CV o o F V C C C G F V G C 0.025 0.30 Average Average Cq=24.25 Cq=20.36 Cyp3a11 Cyp3a11 a" 0.25 a 0.020 0.20 0.015 0.15 0.010 0.10 a" a a" b c b" 0.005 0.05 c b" 0.00 0.000 ll 33 rrooGF 3 GGFFGF+ oll LLCV+ LL3GF+ ttrroCV tt eexx CV oonn VV VVSSVSL3 oonn FFVVSS VSL3 C C C C GG CC GFF CCVV G mRNA (% of 18S rRNA) Cyp3a41a/b a" 0.25 mRNA 18S rRNA) rRNA) mRNA (% (% of of 18S Average Cq=20.29 0.30 mRNA (% of 18S rRNA) mRNA (% of 18S rRNA) Regulation of the Cyp3a Gene Cluster by VSL3 and Conventionalization 0.04 Average Cq=22.85 Cyp3a59 Cyp3a25/59 a" 0.03 ab" c" 0.02 b" 0.01 b" 0.00 C l 3 ro 3 G GF+ F ol LCV+ trCV t GF LGF+ ex CV on V VSVSL3 on F VSVSL3 C C G C GF V Chr 5 Cyp3a57( Cyp3a16( Cyp3a41a( mRNA (% of 18S rRNA) (Cyp3a57 average Cq>30) 0.0012 0.0010 Cyp3a44( Cyp3a11( Cyp3a59( Cyp3a25( Cyp3a41a/b( Average Cq=29.14 Cyp3a16 b" 0.0008 0.0006 0.0004 0.0002 a" a" a" a" 0.0000 C l 3 rGF 3 GGF+ F ol LCV+ LGF+ trCV to ex CV on V VSVSL3 on F VSVSL3 C C G C GF V Selwyn, Cheng, Klaassen, and Cui JY*, 2016 Drug Metab Dispos, 44: 262-74 American College of Toxicology Webinar series Slide 9 Regulation of the Cyp4a Gene Cluster by VSL3 and Conventionalization Selwyn, Cheng, Klaassen, and Cui JY*, 2016 Drug Metab Dispos, 44: 262-74 American College of Toxicology Webinar series Slide 10 Validation of the Protein Expression of Cyp3a and Cyp4a Selwyn, Cheng, Klaassen, and Cui JY*, 2016 Drug Metab Dispos, 44: 262-74 American College of Toxicology Webinar series Slide 11 Validation of Enzyme Activity of Cyp3a and Cyp4a Cyp3a11 Cyp4a14 Selwyn, Cheng, Klaassen, and Cui JY*, 2016 Drug Metab Dispos, 44: 262-74 American College of Toxicology Webinar series Slide 12 PXR- and PPARα-DNA Binding Sites in Control Mouse Liver PXR binding fold-enrichment to Cyp3a loci in mouse liver A Site 5 Site 4 Site 3 Site 2 Site 1 (Data re-analyzed from the PXR ChIP-Seq experiments in control mouse liver, from a previous study by Cui et al., 2009) Cyp3a59' Cyp3a11' Cyp3a44' Cyp3a25' B PPARα-binding to Cyp4a loci in liver Site 6 Site 9 Site Site510 Site Site Site 3 Site 4 8 7 Site 2 Site 1 (Re$analyzed+from+NCBI+GEO+Database+Query+dataset+GSE61817,+sample+ ID+GSM1514929;+original+experiments+were+performed+by+Lee+et+al,+2014.)+ Selwyn, Cheng, Klaassen, and Cui JY*, 2016 Drug Metab Dispos, 44: 262-74 American College of Toxicology Webinar series Slide 13 ChIP of PXR and PPARα binding to Cyp3a and Cyp4a loci Selwyn, Cheng, Klaassen, and Cui JY*, 2016 Drug Metab Dispos, 44: 262-74 American College of Toxicology Webinar series Slide 14 Germ-free and conventionalization have the most prominent effect on the regulation of hepatic drug-processing genes • Germ-free conditions resulted in the most prominent changes in hepatic drug-metabolizing enzyme expression, most notably a consistent down-regulation of many genes in the Cyp3a cluster, but a consistent upregulation of many genes in the Cyp4a cluster. • Conventionalization of GF mice at least partially restores the expression of these genes to CV levels. • The GF and conventionalization mediated changes in Cyp3a and 4a genes are associated with altered PXR and PPARα-binding to the targeted DNA sequences within these genes. American College of Toxicology Webinar series Slide 15 Polybrominated Diphenyl Ethers (PBDEs) • PBDEs are among the most abundant and persistent environmental chemicals in the human population BDE-47 (2,2’,4,4’-Tetrabromodiphenyl ether) BDE-99 (2,2’,4,4’,5-Pentabromodiphenyl ether) • BDE-47 and BDE-99 are the predominant congeners detected in humans, and they are highly prevalent in seafood and breast milk at worrisome levels humans American College of Toxicology Webinar series Slide 16 Hepatic Biotransformation of PBDEs and Toxicities • Hepatic uptake: via organic anion transporting polypeptides (OATP) transporters • Hepatic Phase-I metabolism: by human CYP2B6 to toxic hydroxylated metabolites • Hepatic Phase-II conjugation (in vitro evidence): glucuronidation, sulfonation or methylation • Toxicities of PBDEs: thyroid toxicity, neurodevelopmental disorders, hepatic oxidative stress and cancer. American College of Toxicology Webinar series Slide 17 BDE-47 is a CAR activator Sueyoshi et al., 2014, Toxicol Sci, 137: 292-302 American College of Toxicology Webinar series Slide 18 Sulfotransferase 5a1: important for Phase-II Sulfonation conjugation reaction of xenobiotics • Sult5a1 is up-regulated by the CAR ligand TCPOBOP in livers of wild-type mice • The TCPOBOP-mediated upregulation of Sult5a1 is absent in livers of CAR-null mice • Sult5a1 is a bona fide CARtarget gene • PBDE-47 mediated upregulation of Sult5a1 depends on the presence of gut microbiome. Davis Cowles and Julia Cui American College of Toxicology Webinar series Slide 19 Experimental Design • CV and GF mice were treated with vehicle (corn oil), BDE-47, or BDE-99 via oral gavage once daily for 4-days • GC-MS quantification of PBDE metabolites • RNA-Seq of hepatic transcriptiome American College of Toxicology Webinar series Slide 20 Hepatic Drug-processing Genes Related to PBDE Biotransformation American College of Toxicology Webinar series Cindy Li and Julia Cui Slide 21 PBDE-mediated regulation of the hepatic transcriptome is profoundly modified by lack of gut microbiota • Gut microbiota-dependent regulation • Potentiation effect due to lack of gut microbiota Control vs. BDE-47 GF CV 966 116 754 Control vs. BDE-99 GF CV 629 586 4302 Cindy Li and Julia Cui American College of Toxicology Webinar series Slide 22 Summary of PBDE Oxidative Metabolism in Mouse Liver Cindy Li and Julia Cui in Collaboration with Dr. Irvin Schultz American College of Toxicology Webinar series Slide 23 Overall Conclusion • Gut microbiota regulates the expression and activity of various drug-processing genes in liver • PXR and PPARα appear to be the targets which the intestinal bacteria interact with • The presence of gut microbiota alters the hepatic metabolism of PBDEs, and it is necessary in the PBDEmediated regulation of many hepatic genes involved in drug metabolism and other critical endogenous pathways • Future studies will determine which specific bacterial strains and their microbial metabolites interact with the host receptors in liver American College of Toxicology Webinar series Slide 24 Looking into the Future: Microbiome is a Key Player in Exposome • Microbiome and Toxicology (mice as a research model) • Microbiome and nutrition • Microbiome and early life exposure/epigenetics • Microbiome and “Big Data” • Microbiome as biomarkers for risk assessment • Probiotics as preventive therapy to reduce toxic exposure in vulnerable populations American College of Toxicology Webinar series Slide 25 Acknowledgements Members in the Cui Laboratory Collaborators: • Felcy Selwyn, former Research Scientist • Dr. Bhagwat Prasad (UW) • Dr. Irvin Schultz (PNNL) • Sunny Cheng, Research Scientist • SooWan Lee, Research Scientist • Cindy Li, PhD Student • Joseph Dempsey, PhD Student • Gurkirat Sidhu, Undergraduate Student • Daniel Park, Undergraduate Student • Khakkhak Khayi, Undergraduate Student • Felcy Selwyn, former Research Scientist • Yubin Song, former Undergraduate Student • Shinhee Park, former Undergraduate Student • Elaine Chen, former Undergraduate Student • Davis Houston Cowles, former EHREP Intern Supported by National Institute of Health R01 grants GM111381, ES025708, and R01 ES019487, UW start-up funds, and Murphy Endowment. American College of Toxicology Webinar series Slide 26 Aaron Ericsson, DVM, PhD Dr. Ericsson received a DVM from the University of Missouri (MU) in 2006 and went on to complete a residency in Laboratory Animal Medicine in 2009, followed by a T32-funded PhD in Area Pathobiology, focusing on the role of the innate immune system in gastrointestinal inflammation and colitis-associated colorectal cancer (CAC). In 2013, he assumed the role of lead scientist for microbiome research at both the NIH-funded MU Mutant Mouse Resource and Research Center (MMRRC) and Rat Resource and Research Center (RRRC), and received funding to found the MU Metagenomics Center in 2014. He is primarily funded by a K01 to investigate the influence of certain microbes and polymicrobial communities on CAC, and also collaborates heavily with NIHand USDA-funded investigators. American College of Toxicology Webinar series Slide 27 Julia Yue Cui, PhD Julia received her B.S. Degree in Chukechen Honors College, Zhejiang University in Hangzhou, China, and received her Ph.D. Degree with honors in University of Kansas Medical Center. Julia currently is an Assistant Professor in Toxicology in the Department of Environmental and Occupational Health Sciences. She is a recipient of the Sheldon D. Murphy Endowed Chair, and a member of Center of Ecogenetics & Environmental Health. Julia is trained as a toxicologist, specializing in using toxicogenomic and toxicoepigenomic approaches to determine the effects of environmental chemical exposure and reprogramming the gut microbiome on the transcriptional and epigenetic regulation of genes involved in drug metabolism and obesity during development. American College of Toxicology Webinar series Slide 28 Thank you for your participation in the American College of Toxicology Webinar! We hope to see you at the 37th Annual Meeting of the American College of Toxicology. American College of Toxicology Webinar series Slide 29