* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download From Restriction Maps to Cladograms
Gene expression profiling wikipedia , lookup
Molecular cloning wikipedia , lookup
Pathogenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Minimal genome wikipedia , lookup
Non-coding DNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome (book) wikipedia , lookup
Metagenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Human–animal hybrid wikipedia , lookup
Point mutation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome evolution wikipedia , lookup
DNA barcoding wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Helitron (biology) wikipedia , lookup
Designer baby wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome editing wikipedia , lookup
Koinophilia wikipedia , lookup
History of genetic engineering wikipedia , lookup
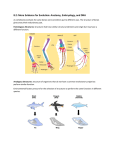
From Restriction Maps to Cladograms Name____________________ Restriction Maps: A particular restriction nuclease will cut DNA into a series of fragments called restriction fragments. If you treat DNA with a number of restriction nucleases, you can construct a map that shows the location of each of these cuts on a given length of DNA. Instructions: Below are restriction maps for a segment of DNA common to a number of mammals including humans: the genes which code for hemoglobin. 1.Compare the restriction map for each species to the human map. Make a mark on the map for each difference. A difference is the addition or subtraction of a restriction site. (See the example). 2.For each restriction map, count the number of differences that you find. Enter these data on a data chart. 3.In the cladogram, enter the number of differences between each species and human. The species with the least number of differences should be entered closest to the human, and the species with the greatest number of differences, farthest from the human. Example: In the example below, Species 2 has three differences with Species 1. Restriction Map of Human and Various Primate DNA in a Cluster of Genes Coding for Hemoglobin Elizabeth Zimmer and Alan Wilson developed the map reproduced below (as cited in Alberts, et al. Molecular Biology of the Cell. Garland Publishing, 1983). From the following list, predict which are the most and which are the least closely related: human, chimp, pygmy chimp, gorilla, orangutan, and gibbon. Cladogram: similarities of restriction maps between humans and other primates. Questions 1.Which species has the most differences with humans? 2.Which species has the fewest differences with humans? 3. Hemoglobin is the molecule in the bloodstream which transfers oxygen from the lungs to the body's cells; it is, therefore, important that it work properly. Is an animal born with large changes in its hemoglobin likely to survive to produce offspring? Reason? 4. Some small changes in the hemoglobin molecule may give an animal an advantage in one environment and a disadvantage in some other environment. Are these sorts of changes likely to accumulate in the hemoglobin gene over long periods of time, or be changed back and forth as the outside environment changes? Explain. 5. Some small changes in the hemoglobin molecule are selectively neutral--they do not affect the animal's fitness in any change of environment. Are these sorts of changes likely to accumulate in the hemoglobin gene over long periods of time, or be changed back and forth as the outside environment changes? Why? 6.Of the three sorts of change introduced in questions 3 - 5, which sort is the most likely to accumulate in the hemoglobin gene from generation to generation? 7. If the changes we are counting are a) random, b) selectively neutral, and c) tend to persist in the genes from generation to generation, would you expect the genes of two closely related organisms to have more differences or fewer differences than the genes of two distantly related organisms?