* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PDF
Microevolution wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epitranscriptome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Minimal genome wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Primary transcript wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression profiling wikipedia , lookup
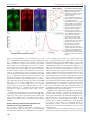
© 2014. Published by The Company of Biologists Ltd | Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 RESEARCH ARTICLE Spatial gradients of protein-level time delays set the pace of the traveling segmentation clock waves ABSTRACT The vertebrate segmentation clock is a gene expression oscillator controlling rhythmic segmentation of the vertebral column during embryonic development. The period of oscillations becomes longer as cells are displaced along the posterior to anterior axis, which results in traveling waves of clock gene expression sweeping in the unsegmented tissue. Although various hypotheses necessitating the inclusion of additional regulatory genes into the core clock network at different spatial locations have been proposed, the mechanism underlying traveling waves has remained elusive. Here, we combined molecularlevel computational modeling and quantitative experimentation to solve this puzzle. Our model predicts the existence of an increasing gradient of gene expression time delays along the posterior to anterior direction to recapitulate spatiotemporal profiles of the traveling segmentation clock waves in different genetic backgrounds in zebrafish. We validated this prediction by measuring an increased time delay of oscillatory Her1 protein production along the unsegmented tissue. Our results refuted the need for spatial expansion of the core feedback loop to explain the occurrence of traveling waves. Spatial regulation of gene expression time delays is a novel way of creating dynamic patterns; this is the first report demonstrating such a control mechanism in any tissue and future investigations will explore the presence of analogous examples in other biological systems. KEY WORDS: Segmentation clock, Oscillation, Traveling wave, Systems biology, Computational modeling, Gene expression INTRODUCTION Vertebral segments differentiate from their embryonic rudiments called somite segments. Segmentation of somites, namely somitogenesis, occurs at a species-specific pace during embryonic development. Somitogenesis is controlled by a gene expression oscillator called the vertebrate segmentation clock that ‘ticks’ in cells located in the unsegmented presomitic mesoderm (PSM). The period of the segmentation clock in cells located at the posterior end of the PSM is equivalent to the period of somite formation. The period of oscillation increases as cells are passively displaced closer to the anterior end of the PSM. The oscillations finally halt as cells bud off as a somite segment at the anterior end of the tissue. Cells exiting the PSM are arrested in different states of the oscillation cycle, defining which portion of a 1 Department of Mathematics, Colgate University, Hamilton, NY 13346, USA. Department of Biology, Colgate University, Hamilton, NY 13346, USA. Department of Computer Science, Colgate University, Hamilton, NY 13346, USA. 4 Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA. *These authors contributed equally to this work 2 3 ‡ Authors for correspondence ([email protected]; [email protected]) Received 26 April 2014; Accepted 30 August 2014 4158 somite they will form and which set of genes they will express during differentiation (Pourquié, 2011). The spatiotemporal dynamics of oscillations create a phase difference among cells along the PSM. Anterior PSM cells are retarded in the oscillator phase relative to posterior PSM cells; the further anterior the cells are located, the greater the retardation is. This phase difference results in the formation of traveling wave-like gene expression patterns along the PSM (Fig. 1A), which can be visualized by in situ hybridization for oscillating mRNAs or realtime imaging of oscillating proteins. The different phases of the oscillator cycle are mapped out in space as stripes of expression of the oscillating genes. The number of expression stripes reflects the number of oscillation cycles by which the anterior cells are lagging behind posterior cells (Giudicelli et al., 2007; Gomez et al., 2008). The spatial period profile of the segmentation clock was measured in zebrafish by utilizing fluorescent in situ hybridization data (Giudicelli et al., 2007). We have further demonstrated that the period profile is conserved between zebrafish and corn snake (Gomez et al., 2008). The prevailing model in the somitogenesis field states that the pacemaker mechanism of the segmentation clock is based on an autoinhibitory transcriptional feedback loop of Her/Hes family genes (Harima et al., 2013; Jensen et al., 2003; Lewis, 2003; Monk, 2003; Özbudak and Pourquie, 2008). Quantitative data have become more available in the somitogenesis field in recent years. Molecular-level models coupled with experimentation have provided insight into how the dynamics of the segmentation clock in the posterior PSM cells can be affected in various mutant conditions (Ay et al., 2013; Hanisch et al., 2013; Özbudak and Lewis, 2008; Schroter et al., 2012). Although we have a good understanding of how the oscillator functions in the posterior PSM, the underlying mechanism that generates the spatiotemporal traveling waves in the whole PSM has remained elusive. A few modeling studies addressing the formation of traveling waves have been carried out (Campanelli and Gedeon, 2010; Cinquin, 2007; Uriu et al., 2009). Although these studies provided insight into the system, these models were based on regulatory assumptions that have been corrected by recent experimental results (Hanisch et al., 2013; Schroter et al., 2012; Trofka et al., 2012). Furthermore, these models focused on simulating the system only in a wild-type background and were therefore largely unconstrained. More rigorous models can be built by constraining them to explain the spatiotemporal dynamics of the segmentation clock in all available genetic backgrounds. Four broad hypotheses could explain the traveling wave-like gene expression patterns in the tissue. The first three hypotheses require the inclusion of new genes into the core clock regulatory network, whereas the fourth does not have such a restriction: (1) instructive regulation of clock gene expression by upstream transcription factors [such as tbx6 (also known as fss and tbx24)] (Brend and DEVELOPMENT Ahmet Ay1,2,‡, Jack Holland3, *, Adriana Sperlea3, *, Gnanapackiam Sheela Devakanmalai4, *, Stephan Knierer4, *, Sebastian Sangervasi1, *, Angel Stevenson4 and Ertuğ rul M. Ö zbudak4,‡ RESEARCH ARTICLE Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 Holley, 2009; Gajewski et al., 2003; Holley, 2007; Holley et al., 2000); (2) inclusion of regionally expressed oscillatory Her family genes into the core clock network (Cinquin, 2007; Schroter et al., 2012; Shankaran et al., 2007); (3) inclusion of anterior PSMspecific genes into the core clock network (Holley, 2007); and (4) spatial variation of biochemical reaction rates of core clock genes (Uriu et al., 2009). Here, we built a molecular-level multicellular model to assess the feasibility of the fourth scenario in generating the traveling waves throughout the PSM. Our model is able to explain the formation of traveling waves in wild-type background as in Giudicelli et al. (2007). The model predicts an increasing gradient of effective translational time delay along the posterior to anterior direction and in this way recapitulates the spatiotemporal profile of the traveling segmentation clock waves in six genetic backgrounds (wild type, her1−/−, her7−/−, hes6−/−, her7−/−;hes6−/− and notch1a−/−). We experimentally measured the effective Her1 translational time delay, which includes translation and nuclear import of proteins, and found that it increases approximately 4-fold towards the anterior PSM in zebrafish embryos and confirmed the major prediction of our model. Our model further predicts how the spatial profiles of period, amplitude and degree of synchronization of the segmentation clock vary in all available genetic backgrounds and lays the foundation for future experimental tests. segmentation clock is yet to be discovered. We reasoned that these potential regulators should change the ‘effective’ rates of selected biochemical reactions along the PSM, without changing the core clock regulatory network, to set the gradient-like period profile along the PSM. To determine which reaction rates influence the period most, we extended our published model (Ay et al., 2013) into 6×10 cells representing the posteriormost tailbud region. Then, we identified parameter sets that robustly reproduced roughly 20 different experimental phenotypes in six different genetic backgrounds as before (Ay et al., 2013). We then performed a sensitivity analysis at different positions in the PSM to determine the reaction parameters whose perturbation influences the oscillation period the most (Fig. 2A). We found that the most influential parameters are time delays in the transcription or translation of the her1, her7 or deltaC genes, the halflives of their three respective mRNAs, and the halflives of repressor dimers (Fig. 2A). These results echoed the single-cell analytic solution of Lewis (2003). However, the sensitivity analysis showed that expression time delays of the deltaC gene also influence the clock period, in agreement with Herrgen et al. (2010). A further difference from Lewis (2003) is that halflives of dimer proteins more strongly affect the period than halflives of monomer proteins (supplementary material Tables S2 and S3). RESULTS Key biochemical reaction rates influencing the period are determined by sensitivity analysis We sought to determine the reaction parameters whose spatial variation could reproduce the measured period profile in wild-type zebrafish embryos (Giudicelli et al., 2007). We applied increasing or decreasing gradients to each parameter along the PSM (6×50 cells), similar to Uriu et al. (2009), and assessed whether gradients of any combinations of influential parameters could match the period profile with a maximum of 10% error. Our results showed that, contrary to an earlier model (Uriu et al., 2009), the gradients in The activities of three signaling pathways generate gradients along the PSM (Pourquie, 2011). Furthermore, many transcription factors are expressed along the PSM regionally that could potentially influence the period of the segmentation clock (Özbudak et al., 2010). How all these potential regulators join forces to generate traveling waves of the Traveling segmentation clock waves can be generated only if the effective time delays increase along the PSM 4159 DEVELOPMENT Fig. 1. The traveling segmentation clock waves. (A) In one oscillation cycle, the expression profiles ( purple corresponds to higher levels of her1 mRNA) return back to the original phase, and one somite segments at the anterior end of the PSM. Oscillations of neighboring cells are locally synchronized by Notch signaling. (B) The regulatory interactions in the zebrafish segmentation clock network. Her1, Her7 and Hes6 proteins can form homo- and heterodimers. However, only the Her1-Her1 homodimer and the Her7-Hes6 heterodimer repress transcription of her1, her7 and deltaC. Notch signaling activates transcription of her1 and her7. RESEARCH ARTICLE Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 increasing gradients of transcriptional time delays of her1+her7 or deltaC or her1+her7+deltaC mRNAs, and translational time delays of Her1+Her7 or DeltaC or Her1+Her7+DeltaC proteins along the PSM (Fig. 2B). Surprisingly, we noticed that a joint increase in the transcriptional and translational time delays of her1 and her7 mRNA or protein could slow down oscillations sufficiently; however, the synchrony in oscillation phases of nearest neighbors is then lost in the anterior PSM (supplementary material Fig. S3). This is due to the creation of an imbalance between the time delays of Her genes functioning intracellularly and that of deltaC functioning intercellularly (see Discussion). Of the remaining four different parameter gradients, the triple transcriptional time delay and triple translational time delay work with more parameter sets compared with time delays in expression of the deltaC gene alone (Fig. 2B). The triple time delays provide robustness into the system. This again reflects the need for balance between the timings of the intracellular and intercellular feedback loops. Fig. 2. Sensitivity analysis. (A) The period of the segmentation clock is sensitive to eight model parameters: transcriptional time delays of deltaC (delaymd), her1 (delaymh1) and her7 (delaymh7); translational time delays of DeltaC (delaypd) and Her1 (delayph1); and degradation rates of her1 (mdh1), her7 (mdh7) and Her1-Her1 (ddgh1h1). (B) Eight sensitive parameters are used to create ten parameter groups to be varied from posterior to anterior. The gradients of six out of ten parameter groups resulted in similar period behavior to previously measured oscillation period data (Giudicelli et al., 2007). The distributions of successful numbers of parameter sets (out of 165 tested) for varying gradients are shown. Triple transcriptional and translational time delays lead to the highest frequency of parameter sets satisfying the period condition. halflives of mRNAs or proteins do not slow down the period sufficiently along the PSM and cannot accommodate the formation of two expression stripes. Jointly decreasing gradients of degradation rates of the two Her genes or of all three oscillating genes did not change the outcome. These modifications rather dampened the oscillations in the intermediate PSM (supplementary material Fig. S1). Furthermore, we experimentally measured the stability of Her7 protein at different locations along the PSM (see Materials and Methods). We induced a short pulse of Her7 expression in transgenic embryos (Giudicelli et al., 2007) and observed that Her7 protein degrades uniformly along the PSM (supplementary material Fig. S2). This result rules out a regionalized protein degradation mechanism for generating traveling waves and confirms the conclusion of our model. Among all parameter combinations tested, however, there were six that slowed down the period in a manner similar to that measured in wild-type zebrafish embryos (Giudicelli et al., 2007). These are 4160 We next simulated the spatiotemporal dynamics of the segmentation clock in wild-type and five mutant backgrounds using four successful parameter gradients. For each gradient, the rate changes that give the highest number of successful parameter sets that fit to the period observations in wild-type embryos, together with the corresponding parameter sets, are selected for anterior PSM simulations (Fig. 2). Our model successfully reproduces the formation of distinct expression waves in wild-type, hes6−/− and her7−/−;hes6−/− backgrounds (Fig. 3; supplementary material Figs S4-S6 and Movies 1-6). The snapshots resemble published in situ hybridization images in these genetic backgrounds (Fig. 3A) (Schroter et al., 2012). We and others have recently reported that her1−/− embryos halt their oscillations earlier in the PSM and manifest only one or two expression waves at mid-somitogenesis stages; this results in the formation of one less stripe in the anterior PSM in her1−/− mutants (Choorapoikayil et al., 2012; Hanisch et al., 2013) compared with wild-type, hes6−/− and her7−/−;hes6−/− embryos (Schroter et al., 2012). Our model reproduced this observation successfully (Fig. 3). We and others have also reported that traveling wave-like expression patterns are significantly disturbed in her7−/− mutants; there is also a large variation from embryo to embryo from the same mutant clutch (Choorapoikayil et al., 2012; Hanisch et al., 2013; Schroter et al., 2012). Our model nicely recapitulates this phenotype. Snapshots of the simulation movie show that, within a given embryo, the traveling wave pattern is disturbed significantly and the disturbance varies from phase to phase, reflecting the variation observed in embryos from the same clutch (Fig. 3). In the Notch pathway mutants, oscillations are desynchronized and the traveling wave pattern is disturbed. Our simulations readily reproduce this phenotype. The synchronized traveling wave pattern of wild-type embryos is lost in notch1a−/− mutants and instead a salt-and-pepper-like expression profile is observed at each snapshot (Fig. 3). Spatial profiles of the period, amplitude and synchronization scores of the segmentation clock in all genetic backgrounds are calculated from simulations We measured how the period, amplitude and synchronization of the segmentation clock vary in cells located at different positions along DEVELOPMENT The model successfully reproduces spatiotemporal expression of the segmentation clock in wild-type and mutant backgrounds RESEARCH ARTICLE Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 Fig. 3. her1 mRNA levels in the whole PSM in different genetic backgrounds under the influence of a Her1-Her7-DeltaC translational time delay gradient. (A) Snapshots of 6×50 cells located along the PSM in different genetic backgrounds. Images are obtained by setting an increasing gradient to the translational time delay of Her1, Her7 and DeltaC proteins. Traveling waves are readily generated under these conditions. Darker color corresponds to higher levels of her1 mRNA. (B) her1 mRNA levels in six cells for the 300 min have been plotted as they travel from the posterior to anterior PSM in different genetic backgrounds. Period and amplitude increase in all genetic backgrounds. Early arrest of oscillations in the her1−/− mutant and loss of synchronization in her7−/− and notch1a−/− mutants are observed. The increased gradient of effective translational time delays of segmentation clock genes underlies the formation of traveling waves along the PSM Our model outlined two potential mechanisms to generate synchronized traveling waves in the PSM: an increasing gradient of time delays of the transcription or of the translation of oscillating segmentation clock genes along the PSM (Figs 2–5; supplementary material Figs S4-S12). Which one of these two possibilities is utilized to generate the characteristic spatiotemporal dynamics of the segmentation clock? We previously measured the transcriptional time delays of her1, her7 and deltaC genes to be approximately 10-12 min in the anterior PSM (Giudicelli et al., 2007; Hanisch et al., 2013). However, our model utilizes the same parameter range for cells located in the posterior PSM to reproduce phenotypes in all genetic backgrounds. Therefore, the experimentally measured rates in anterior PSM cells exactly correspond to those rates necessitated by our model for posterior PSM cells. These results argue against the possibility of a significant increase in transcriptional time delays along the PSM. To determine the spatial variation of the effective protein production time delay of oscillating genes, we experimentally measured this rate for Her1 protein by utilizing a transgenic animal 4161 DEVELOPMENT the PSM (Fig. 4; supplementary material Figs S7-S9). The parameter sets were selected to match to the Giudicelli et al. (2007) data in the 0% to 80% length of PSM; therefore, the period profiles in the wild-type background show the expected increase along the PSM. Strikingly, the period profiles in individual cells in all mutants also present similar increases along the PSM independently of whether synchronized traveling waves can be observed at the tissue level (Fig. 4A). This prediction of the model may be tested following the future development of more sensitive real-time reporters that allow imaging at the posterior PSM (Delaune et al., 2012). The amplitude of oscillations in our model mildly increases along the PSM (Fig. 4B). Such an increase had previously been observed experimentally at the level of deltaC mRNA (Jiang et al., 2000) and Her1 protein (Delaune et al., 2012). The synchronization scores reveal near-perfect synchrony in wild type, hes6−/−, her1−/− and her7−/−;hes6−/− mutants, reproducing the synchronized traveling waves in these backgrounds (Fig. 4C). The synchronization score of notch1a−/− mutants is close to zero, reflecting the absence of cellcoupling in this background. The synchronization score of her7−/− mutants is very low in the posterior PSM, echoing our previous results (Ay et al., 2013) (Fig. 4C). RESEARCH ARTICLE Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 of mRNA stripes lies further anteriorly than that of protein stripes, i.e. while cells located anteriorly have just started to produce mRNAs, those located posteriorly have already past this phase of their oscillation cycle and started to make proteins. We measured the spatial distance between the onset of cytoplasmic mRNA and nuclear protein stripes. This distance is used to calculate the effective Her1 translational time delay (see Materials and Methods). To our initial surprise, we observed that the effective translational time delay increases 4.4-fold from the intermediate to the anterior PSM in zebrafish embryos (Fig. 6; supplementary material Figs S13-S21). DeltaC translational time delays have previously been measured as approximately 30 min in anterior PSM (Giudicelli et al., 2007). In our simulations, the DeltaC translational time delay has been estimated to be approximately 12 min to satisfy all mutant phenotypes in posterior PSM. This constitutes a 2.5-fold increase in the translational time delay of DeltaC protein from posterior PSM to anterior PSM. We have now measured Her1 translational time delays to have increased approximately 4.4-fold in the anterior PSM. We performed new simulations to confirm that non-symmetric increases in the translational time delays also recapitulate the synchronized traveling waves in the PSM. We fixed the translational time delay of DeltaC protein to a 2.5-fold increase from posterior to anterior PSM, and searched for all the possible Her1 and Her7 translational time delay gradients that could still match all the available experimental observations for the segmentation network. We found that a 4- to 9-fold increase in Her1 and Her7 translational time delays in the PSM reproduce the experimentally observed increase in period (Giudicelli et al., 2007) (Fig. 6F; supplementary material Figs S14-S16). This result agrees well with our experimental observations of an increased Her1 translational time delay in the anterior PSM. Fig. 4. Spatial profiling of period, amplitude and synchronization scores along the PSM in all genetic backgrounds under the influence of a Her1Her7-DeltaC translational time delay gradient. (A,B) The period (A) and amplitude (B) of oscillations are increased as cells traverse from the posterior to the anterior PSM in all genetic backgrounds. (C) The oscillations are locally synchronized in wild type, her1−/−, hes6−/− and her7−/−;hes6−/− mutants. The synchrony is lost in her7−/− and notch1a−/− mutants; however, the synchrony in her7−/− mutants is increased in the anterior PSM. (A-C) x-axis corresponds to spatial location in the PSM; y-axis corresponds to the period, amplitude or synchrony score, which are normalized to corresponding wild-type values in the posterior PSM. that expresses Her1-Venus fusion protein (Delaune et al., 2012). We simultaneously detected the spatial profiles of her1-Venus mRNA and protein by fluorescent in situ hybridization and immunohistochemistry along the PSM. The slowdown of oscillations towards the anterior PSM causes a phase delay in cells located in the anterior PSM compared with those located in the posterior PSM. Owing to this phase difference, the anterior domain 4162 We first performed simulations for cells located in the posterior PSM and trained the model with roughly 20 different phenotypes obtained in six genetic backgrounds. This narrowed down the parameter space that can generate all observed phenotypes simultaneously. Furthermore, we trained our model by matching the predicted rate of slowdown to the rate experimentally observed in zebrafish and by requiring the occurrence of two to three traveling waves in the PSM. Finally, we utilized recently updated information on the nature of functional repressor dimers in zebrafish. All these updates collectively reduced the set of parameters whose variation along the PSM could account for the observed traveling waves; and these parameters can be experimentally measured. The results of our simulations argue against previous claims that the traveling waves are due to a gradient of hes6 transcription along the PSM (Cinquin, 2007) or gradients of effective halflives of mRNA and proteins along the PSM (Uriu et al., 2009). Our analysis shows, moreover, that the traveling waves can be generated without resorting to assumptions of faster decay of monomers versus dimers and an order of magnitude longer halflife of hes6 versus her7 (Campanelli and Gedeon, 2010). Uriu et al. (2009) listed slower mRNA transcription and Delta protein synthesis in posterior than in anterior PSM among the possible scenarios explaining the formation of traveling waves in the PSM. These ideas run counter to our model, which posits that increased, rather than decreased, time delays in transcription and translation along the posterior-anterior direction slow down the oscillations and result in traveling waves. DEVELOPMENT DISCUSSION Constraining mathematical models with quantitative experimental data restricts the number of potential mechanisms generating traveling waves in the PSM RESEARCH ARTICLE Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 The time delays in the expression of Her genes that constitute the intracellular feedback loop and the deltaC gene that forms the intercellular feedback loop have to be balanced (Herrgen et al., 2010; Lewis, 2003). Skewing the relationship of this balance could result in desynchronized oscillations of neighboring cells. Therefore, the model was not able to reproduce synchronized oscillations along the PSM by only increasing the time delays of Her genes (supplementary material Fig. S3). The model shows that increasing the time delays of three oscillating genes along the PSM is a more robust mechanism of generating traveling waves in the tissue (Fig. 2B). Spatiotemporal profiles of the segmentation clock in various genetic backgrounds are readily satisfied with the model The parameter sets that satisfy the requirement of formation of two to three expression waves in the simulations for wild-type embryos also reproduce the in situ hybridization profiles that were published in various mutant conditions (Choorapoikayil et al., 2012; Hanisch et al., 2013; Jiang et al., 2000; Schroter et al., 2012). Two to three synchronized traveling waves are obtained in hes6−/− and her7−/−; hes6−/− mutants as in wild-type embryos (Fig. 3). The simulations demonstrate formation of one less stripe and the arrest of oscillations in more posterior locations in her1−/− mutants than in wild-type embryos (Fig. 3), again matching previous observations. However, the oscillations arrest at high expression levels in our simulations rather than at the low expression level that is experimentally observed. This, we suggest, reflects the fact that our current model does not account the many additional genes (in addition to her1, her7, hes6 and deltaC) that are expressed in a patterned manner in the anterior PSM and/or define the extent of the PSM as a whole. These additional genes might shut off expression of the clock genes at the end of the anterior PSM. One candidate for such a role is the ripply1 gene: expression of this gene is restricted to the anterior PSM and it is crucial for switching off the expression of segmentation clock genes as cells segment into somites (Kawamura et al., 2005). The synchronization scores of hes6−/−, her7−/−;hes6−/−, her1−/− and wild-type embryos are similar along the tissue (Fig. 4C). There is no synchronization of oscillations in the notch1a−/− mutants 4163 DEVELOPMENT Fig. 5. Density plots of traveling waves in all genetic backgrounds under the influence of a Her1-Her7-DeltaC translational time delay gradient. (A-F) Traveling waves are observed in wild-type, her1−/−, hes6−/− and her7−/−; hes6−/− but are blurred in her7−/− and notch1a−/− embryos. The oscillations in her1−/− mutants arrest at more posterior locations. The x-axis reflects time (min) and the y-axis maps spatial locations in the PSM ( posterior is top and anterior is bottom). RESEARCH ARTICLE Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 (Fig. 4C). The synchronization score of oscillations is very low in her7−/− mutants in the posterior PSM (Fig. 4C), which explains why her7−/− mutants are sensitized and oscillations fail frequently during the formation of posterior somites in many of the mutant embryos. However, the synchronization score of her7−/− mutants recovers appreciably along the PSM, reflecting the formation of irregular stripes in some of the embryos in a clutch of mutants that are fixed at different oscillation phases (Hanisch et al., 2013). This result can be explained by the absence of expression of the hes6 gene in the anterior PSM. Thus, the cells located in the anterior PSM of her7−/− mutants effectively lack expression of both her7 and hes6; these cells resemble their counterparts in the her7−/−;hes6−/− double mutant, with the exception that they experience oscillations with compromised synchrony when they were in the posterior PSM before they join the anterior PSM domain. Therefore, the synchronization score in anterior PSM cells of her7−/− mutants never recovers to the levels of the wild type, and her7−/−;hes6−/− mutants and likewise her7−/− mutants never form stripy expression waves that are as defined and regular as those of her7−/−;hes6−/− embryos. In all the genetic backgrounds that we examined, the amplitudes of oscillations increase towards the anterior end of the PSM. These results align with previous experimental results indicating that levels of deltaC mRNA (Jiang et al., 2000) and Her1 protein (Delaune et al., 2012) increase in a similar way before cells exit the PSM. Spatial variation of protein level time delay induces the traveling waves of clock gene expression Our previously published experimental measurements of transcriptional time delays of three genes in the anterior PSM cells (10-12 min) (Giudicelli et al., 2007; Hanisch et al., 2013) were 4164 as low as postulated by our model for the posterior PSM cells. Hence, the experimental data argue against a drastic increase in the transcriptional time delays along the PSM. However, we previously measured the translational time delay of DeltaC protein in the anterior PSM and found it to be more than 30 min (Giudicelli et al., 2007). The experimentally measured value is similar to that predicted by our model for anterior PSM cells and about 2- to 3-fold greater than that of posterior PSM cells. DeltaC antibody staining is insufficiently sensitive for the measurement of low levels of DeltaC protein in the posterior PSM, so that we cannot experimentally measure the DeltaC translational time delay in posterior regions. Using a transgenic animal that was generated by fusing Venus coding sequences to her1 (Delaune et al., 2012), we now measured the effective translational time delay of Her1 protein and found this to increase 4.4-fold in cells located in anterior versus intermediate PSM. Our simulations suggested that respective increases in the translational time delays of Delta/Her proteins can recapitulate the formation of synchronized traveling waves along the PSM (supplementary material Figs S14-S16). Although we have optimized the parameters in our model for the zebrafish system, the results of our study are general and can be extended to other vertebrate organisms. The transcriptional time delays were first determined in the zebrafish model (Giudicelli et al., 2007; Hanisch et al., 2013). Recently, this technique has been adapted to measure the transcriptional time delays in other vertebrate species (Hoyle and Ish-Horowicz, 2013; Takashima et al., 2011). It now remains to be determined whether the spatiotemporal regulation of effective translational time delays, which has now been discovered in zebrafish, is conserved in other vertebrate species. Alternative models were previously put forward to explain the formation of traveling waves. Expression of clock genes is lost in DEVELOPMENT Fig. 6. Effective Her1 translational time delay increases in the anterior PSM. (A-C) Fluorescent in situ hybridization of Tg(her1:her1-Venus) transgenic embryos with a Venus antisense probe (A), immunohistochemistry against Venus protein (B) and merge of RNA and protein staining with DAPI counterstaining (blue) (C). (D) Spatial profiles of RNA (green) and protein (red) levels from A-C. Solid lines are smoothed profiles. The short arrow indicates d (spatial interval) and the long arrow indicates S (spatial wavelength). (E) Effective Her1 protein production time delay is plotted along normalized PSM length. The measurements are made from intermediate to anterior PSM. The data are binned at equal spatial distances and the mean (±s.e.m.) is plotted along the axis. (F) The number of parameter sets (out of the 165 robust parameter sets satisfying posterior PSM experimental conditions) that reproduce period elongation in the tissue as the effective protein production time delay for Her proteins increases at different times (DeltaC protein production time delay is set to a 2.5-fold increase along the full PSM). the anteriormost PSM in tbx6 mutants (Holley, 2007); distinct cisregulatory elements have been identified to drive expression of the her1 clock gene specifically in the anterior PSM (Brend and Holley, 2009; Gajewski et al., 2003) and these DNA sequences are bound by upstream transcription factors (Brend and Holley, 2009). If the upstream transcription factors create a new negative-feedback loop and their activities oscillate with a long period in the anterior PSM, then they could impose a longer oscillation period on Her/Delta gene expression than the period obtained by the time delays of the Her autoinhibitory feedback loop in the posterior PSM. A second alternative mechanism is the spatially tiled expression of various Her family oscillatory genes in the PSM. Although the expression of master oscillatory genes (her1, her7 and deltaC) exhibit traveling waves in the tissue, oscillations of other Her genes are confined to different spatial locations in the PSM (Shankaran et al., 2007). Integration of regionally expressed Her genes into the core regulatory network has been proposed to underlie positionspecific control of the oscillation period (Schroter et al., 2012; Shankaran et al., 2007). Furthermore, many additional genes are expressed differentially in the anterior PSM (Holley, 2007; Özbudak et al., 2010); these genes could potentially regulate expression of Her/Delta genes in the anterior PSM. All three alternative models necessitate the integration of additional genes into the core clock network to generate the traveling segmentation clock waves. The results of our interdisciplinary approach negate such a need. Even though different factors regulate the expression of oscillating clock genes in the anterior PSM, these regional regulators act permissively for clock gene expression and they do not expand the core negative-feedback loop. The molecular mechanism that controls such a spatiotemporal transition in effective biochemical rates in the segmentation clock is currently unknown. It is likely to involve gradients of signaling pathways and genes that are expressed in the anterior PSM and define its extent and special character. The effective protein-level time delay that we experimentally measured includes the translation and intracellular transport of proteins. Regulation of translational time delay has previously been observed (Buchan and Stansfield, 2007; Lakkaraju et al., 2008). However, the rate of nuclear trafficking contributes significantly to the period of the circadian clock (Meyer et al., 2006; Tataroğlu and Schafmeier, 2010). We previously found that the expression of many genes regulating intracellular protein transport is downregulated in the anterior compared with the posterior PSM (Özbudak et al., 2010). The protein products of some of these genes could modify expression time delays of the segmentation clock genes in the anterior PSM. Future quantitative experiments coupled with computational modeling will reveal the molecular mechanism underlying this spatial transition. MATERIALS AND METHODS Protein time delay measurement Double fluorescent Venus transcript and protein staining was carried out with 12- to 14-somite stage Tg(her1:her1-Venus)bk15 (Delaune et al., 2012) transgenic embryos. Antisense Venus RNA probe was synthesized by in vitro transcription from linearized plasmids using digoxigenin RNA labeling mix (Roche). Embryos were incubated jointly with peroxidaseconjugated anti-digoxigenin (Roche, 11207733910; 1/500) and anti-GFP rabbit IgG fraction (Molecular Probes, A-11122; 1/200) antibodies overnight. The peroxidase activity was detected using the TSA Plus Fluorescein system (PerkinElmer). The peroxidase activity was then quenched with 1% H2O2, followed by extensive washes. Embryos were incubated with secondary peroxidase-conjugated AffiniPure goat anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories, 111-035-144; Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 1/200) and peroxidase activity was detected using the TSA Plus TMR system (PerkinElmer). Specimens were counterstained with DAPI, flatmounted in 50% glycerol, and imaged with a Zeiss Imager Z2 microscope equipped with Apotome and Axiovision 4.8 Rel. Fluorescent images were taken at 1-3 μm serial sections and 2D images were created by projecting all images taken at different focal planes. DAPI staining is used to determine the last segmented somite and the posterior end of the notochord. As Her1 protein is short lived and we carried out stringent double RNA-protein staining, the protein signal can be robustly detected only in the nucleus, where proteins are confined in a much smaller area and the signal is higher than background noise in the fluorescence channel. The image is warped in Adobe Photoshop to make the stripes of gene expression appear at right angles to the body axis, and the notochord is blanked out leaving only PSM tissue to contribute to the analysis, as in Hanisch et al. (2013). The spatial profiles of mRNA and protein staining are plotted using the Analyze:Plot Profile tool of ImageJ. For each point along the axis, we took the total signal summed over the transverse column of pixels (supplementary material Table S4) and used Gaussian smoothing (see supplementary methods for explanation and code) to obtain a smooth graph (Hanisch et al., 2013). By comparing two graphs, we calculated the spatial interval (d) between the onset of cytoplasmic mRNA signal and that of the nuclear protein signal for selected stripes in the anterior and intermediate PSM (Giudicelli et al., 2007; Hanisch et al., 2013). The spatial profile of the oscillation period, T(x), is obtained from Giudicelli et al. (2007). The spatial wavelength, S(x), is calculated by the distance between consecutive peaks or troughs of RNA or protein expression levels. We converted this spatial distance to a time interval using the following equation (Giudicelli et al., 2007): tp(x)=30×d(x)×T(x)/S(x). The time interval corresponds to the effective translational time delay including translation and nuclear import of proteins (supplementary material Table S5). Since calculations necessitate two consecutive expression stripes (Giudicelli et al., 2007), we could not determine the effective translational time delay in the posterior PSM as the signal of protein staining is too weak in that region. Data from a set of 65 embryos are pooled together and binned at equal axis width to determine the spatial profile of proteinlevel time delay in the PSM. Protein degradation measurement Tg(hsp70l:HA-her7) (Giudicelli et al., 2007) transgenic embryos at 12- to 14-somite stages were heat shocked in a water bath for 40 min at 37°C, dechorionated manually, and incubated in embryo media containing 100 μg/ml cycloheximide for 10 min at 28°C. Then, embryos were collected every 5 min, fixed and stained with a rat monoclonal anti-HA antibody (Roche, 3F10; 1/500) in combination with Alexa Fluor 647 goat anti-rat IgG secondary antibody (Life Technologies, A21247; 1/400). Specimens were counterstained with DAPI, imaged as single sections and spatial protein profiles were measured as described above. The PSM was divided into ten equal sections; protein levels were averaged in each section, normalized to the mean of all sections and plotted. Deterministic model We have developed a time-delayed differential equation (DDE) model consisting of 14 equations with 44 parameters (Ay et al., 2013) (see supplementary material Table S1). Our model includes her1, her7, hes6 and deltaC genes. Her1, Her7 and Hes6 proteins form six different homoand heterodimers. However, only Her1-Her1 and Her7-Hes6 dimers are able to repress transcription of the her1, her7, deltaC genes (Fig. 1B). DeltaC ligand binds to a Notch receptor and results in proteolytic cleavage of the intracellular domain of Notch protein (NICD). NICD translocates into the nucleus to activate transcription of her1 and her7. The transcriptional repressors are assumed to compete with the NICD protein for binding to the DNA regulatory region to repress transcription of her1 and her7 (Özbudak and Lewis, 2008). Following Lewis (2003), we do not explicitly write an equation representing the production of NICD. The translational time delay of DeltaC effectively incorporates translation and membrane localization of DeltaC protein, interaction of Delta-Notch proteins, and production and nuclear localization of NICD. The translational time delays of Her1 and Her7 proteins include the translation and nuclear import of repressor 4165 DEVELOPMENT RESEARCH ARTICLE RESEARCH ARTICLE Spatial modeling The PSM tissue is represented as a two-dimensional hexagonal grid of 6×50 cells in our simulations. We artificially connected the right- and leftmost cells in each column, such that each cell has six neighbors, except for the cells located in the most posterior and anterior columns, which have four neighbors. The model is simulated for 1200 min in total: (1) we first simulated only the 6×10 cells forming the posterior PSM for 600 min; (2) to represent the posterior growth of the PSM, we then added a column of six cells every 6 min for the next 240 min until the PSM was full; (3) after the PSM reaches its full size of 6×50 (300) cells, a column of six cells was added at the posterior end and removed at the anterior end of the PSM in the last 360 min, preserving a fixed PSM size. We set the hes6 mRNA synthesis rate (msh6) to a biologically relevant parameter range in the posterior PSM (first ten columns of cells); this rate is linearly interpolated at all intermediate PSM locations (20 columns of cells), and we finally set this synthesis rate to zero in the anterior PSM (last 20 columns of cells). Similarly, we varied time delay and degradation rates suggested by the sensitivity analysis (described below) from the posterior end of the PSM (eleventh column of cells) to the anterior end of the PSM (fiftieth column of cells). The reaction rates were set to the same level (with up to ±5% perturbation) in all cells up to the tenth column and the rates are increased or decreased linearly until a specified level at the end of the anterior PSM. Time delays were increased up to tenfold and degradation rates were decreased down to 1% at the end of anterior PSM. Pseudo-stochastic simulations In order to reflect randomness in biochemical reactions of the segmentation clock network, we carried out pseudo-stochastic simulations of our model. Randomness has been introduced by allowing each cell to have up to 10% difference in rates for each biochemical reaction. The parameter values that have been randomly selected from the biologically relevant ranges have been perturbed up to ±5% for each cell. This extrinsic noise is kept constant throughout the lifetimes of cells in the PSM. The perturbed system has been numerically solved with Euler’s method by incrementing time at every 0.01 min, and updating mRNA and protein levels at each iteration using rate of changes provided by the model. Parameter estimation and sensitivity analysis mRNA and protein decay rates and transcriptional time delays have been measured previously (Ay et al., 2013; Giudicelli et al., 2007; Hanisch et al., 2013). However, some of the reaction rates in the segmentation clock network have not been experimentally measured. In our study, initial parameter sets were composed of randomly generated parameter values within biologically relevant ranges. In the posterior PSM simulations, we employed a global parameter estimation algorithm called stochastic ranking evolutionary strategy (SRES) to identify the parameter sets that give the best model fitness to experimental data in wild type and her1−/−, her7−/−, hes6−/−, her7−/−;hes6−/− and notch1a−/− mutants (supplementary methods). The SRES method creates superior results in large-scale biological systems (Fakhouri et al., 2010; Fomekong-Nanfack et al., 2007; Moles et al., 2003). To test the impact of different reaction rates on the segmentation clock period, we employed sensitivity analysis at different spatial points along the PSM (cell#: 5, 15, 20, 25, 40). The sensitivity analysis was performed using a local sensitivity analysis technique (supplementary methods) at 165 diversely selected robust parameter sets that satisfy the experimental conditions in the posterior PSM. In the anterior PSM simulations, the parameter sets obtained from the posterior simulations are pruned by comparing the simulated period profiles of the her1 mRNA in 30 neighboring cells with the experimental period profile of Giudicelli et al. (2007). For each reaction rate gradient, we selected parameter sets reproducing the experimental period profile (within ±10% 4166 variation) in 80% of the PSM in more than 24 out of 30 cells. These parameter sets and specific gradients were used to create spatial movies and snapshots and to calculate period, amplitude and synchronization profiles. Analysis of oscillation dynamics Posterior PSM The model was run for 600 min in 60 posterior PSM cells, and expression levels of her1 mRNA were used to calculate oscillation features in the posterior PSM. The period was calculated as the time difference between the last two peaks, while amplitude was calculated as the average expression level difference between the last peak and two flanking troughs. The synchronization score was calculated as the average of the Pearson correlation coefficients between each cell and the cell located roughly in the middle of the posterior PSM. Whole PSM After the PSM reached its full size of 300 cells, we followed the next added five columns of cells (30 cells in total) for 300 min (total time that they spent in the PSM). For each cell, the peak and troughs of her1 mRNA oscillations were used to calculate the period and amplitude of oscillations. The period was calculated as the time difference between two consecutive peaks, and the amplitude was calculated as the average expression level change between a peak and two flanking troughs. For a single parameter set, the average of period and amplitude at eight spatial locations (cell#: 5, 12, 17, 22, 27, 32, 37, 42) was calculated. The synchronization score was measured by finding the average of the Pearson correlation coefficients between each cell and the first cell in a given column and this was then averaged over five consecutive columns of cells. For a single parameter set, the average of synchronization levels at nine spatial locations (cell#: 5, 10, 15, 20, 25, 30, 35, 40, 45) was calculated. Coding The codes have been implemented in C++ and Python (codes are available upon request). C++ was used due to its speed and Python was used due to its ease of use in plotting libraries. The current version of our code can perform a 1200-min simulation of 300 PSM cells in less than 1 min (on an iMAC running MAC OSX 10.8.5 with 3.06 GHz Intel Core 2 Duo and 8 GB of RAM). A parallel version of the code was written using the Message Passing Interface (MPI) for time-intensive simulations. Acknowledgements We thank Sharon Amacher and Emilie Delaune for sharing the Tg(her1:her1bk15 transgenic line and protocols; Julian Lewis, Stuart Newman and Priscilla Venus) Van Wynsberghe for critically reading the manuscript; and Spartak Kalinin for fish care. Competing interests The authors declare no competing financial interests. Author contributions A.A. and E.M.O. designed the project, built the mathematical model and analyzed experimental data. A.A., J.H., A. Sperlea and S.S. executed the computational simulations. G.S.D., S.K., A. Stevenson and E.M.O. performed the experiments. A.A., J.H., A. Sperlea, G.S.D., S.K., S.S., A. Stevenson and E.M.O. wrote the manuscript. Funding This study was supported by Colgate University NASC Division funds (to A.A.), by an Alfred P. Sloan Foundation Research Fellowship (to E.M.O.) and by the National Institutes of Health [1 R01 GM111987 01 to E.M.O.]. Deposited in PMC for release after 12 months. Supplementary material Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.111930/-/DC1 References Ay, A., Knierer, S., Sperlea, A., Holland, J. and Ö zbudak, E. M. (2013). Shortlived Her proteins drive robust synchronized oscillations in the zebrafish segmentation clock. Development 140, 3244-3253. DEVELOPMENT proteins, while the transcriptional time delays of her1, her7 and deltaC mRNAs include transcription, splicing and nuclear-to-cytoplasmic export of mRNAs. Each equation describes the rate of change of an mRNA, a monomer protein or a dimer protein. Reaction terms describe the synthesis and degradation of mRNAs and proteins as well as dimer association, dissociation and degradation. Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 Brend, T. and Holley, S. A. (2009). Expression of the oscillating gene her1 is directly regulated by Hairy/Enhancer of Split, T-box, and Suppressor of Hairless proteins in the zebrafish segmentation clock. Dev. Dyn. 238, 2745-2759. Buchan, J. R. and Stansfield, I. (2007). Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell 99, 475-487. Campanelli, M. and Gedeon, T. (2010). Somitogenesis clock-wave initiation requires differential decay and multiple binding sites for clock protein. PLoS Comput. Biol. 6, e1000728. Choorapoikayil, S., Willems, B., Strö hle, P. and Gajewski, M. (2012). Analysis of her1 and her7 mutants reveals a spatio temporal separation of the somite clock module. PLoS ONE 7, e39073. Cinquin, O. (2007). Repressor dimerization in the zebrafish somitogenesis clock. PLoS Comput. Biol. 3, e32. Delaune, E. A., François, P., Shih, N. P. and Amacher, S. L. (2012). Single-cellresolution imaging of the impact of notch signaling and mitosis on segmentation clock dynamics. Dev. Cell 23, 995-1005. Fakhouri, W. D., Ay, A., Sayal, R., Dresch, J., Dayringer, E. and Arnosti, D. N. (2010). Deciphering a transcriptional regulatory code: modeling short-range repression in the Drosophila embryo. Mol. Syst. Biol. 6, 341. Fomekong-Nanfack, Y., Kaandorp, J. A. and Blom, J. (2007). Efficient parameter estimation for spatio-temporal models of pattern formation: case study of Drosophila melanogaster. Bioinformatics 23, 3356-3363. Gajewski, M., Sieger, D., Alt, B., Leve, C., Hans, S., Wolff, C., Rohr, K. B. and Tautz, D. (2003). Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130, 4269-4278. Giudicelli, F., Ö zbudak, E. M., Wright, G. J. and Lewis, J. (2007). Setting the tempo in development: an investigation of the zebrafish somite clock mechanism. PLoS Biol. 5, e150. Gomez, C., Ö zbudak, E. M., Wunderlich, J., Baumann, D., Lewis, J. and Pourquié , O. (2008). Control of segment number in vertebrate embryos. Nature 454, 335-339. Hanisch, A., Holder, M. V., Choorapoikayil, S., Gajewski, M., Ö zbudak, E. M. and Lewis, J. (2013). The elongation rate of RNA Polymerase II in the zebrafish and its significance in the somite segmentation clock. Development 140, 444-453. Harima, Y., Takashima, Y., Ueda, Y., Ohtsuka, T. and Kageyama, R. (2013). Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Rep. 3, 1-7. Herrgen, L., Ares, S., Morelli, L. G., Schrö ter, C., Jü licher, F. and Oates, A. C. (2010). Intercellular coupling regulates the period of the segmentation clock. Curr. Biol. 20, 1244-1253. Holley, S. A. (2007). The genetics and embryology of zebrafish metamerism. Dev. Dyn. 236, 1422-1449. Holley, S. A., Geisler, R. and Nusslein-Volhard, C. (2000). Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev. 14, 1678-1690. Hoyle, N. P. and Ish-Horowicz, D. (2013). Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proc. Natl. Acad. Sci. USA 110, E4316-E4324. Development (2014) 141, 4158-4167 doi:10.1242/dev.111930 Jensen, M. H., Sneppen, K. and Tiana, G. (2003). Sustained oscillations and time delays in gene expression of protein Hes1. FEBS Lett. 541, 176-177. Jiang, Y.-J., Aerne, B. L., Smithers, L., Haddon, C., Ish-Horowicz, D. and Lewis, J. (2000). Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475-479. Kawamura, A., Koshida, S., Hijikata, H., Ohbayashi, A., Kondoh, H. and Takada, S. (2005). Groucho-associated transcriptional repressor ripply1 is required for proper transition from the presomitic mesoderm to somites. Dev. Cell 9, 735-744. Lakkaraju, A. K. K., Mary, C., Scherrer, A., Johnson, A. E. and Strub, K. (2008). SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133, 440-451. Lewis, J. (2003). Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 13, 1398-1408. Meyer, P., Saez, L. and Young, M. W. (2006). PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311, 226-229. Moles, C. G., Mendes, P. and Banga, J. R. (2003). Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res. 13, 2467-2474. Monk, N. A. M. (2003). Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr. Biol. 13, 1409-1413. Ö zbudak, E. M. and Lewis, J. (2008). Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS Genet. 4, e15. Ö zbudak, E. M. and Pourquié , O. (2008). The vertebrate segmentation clock: the tip of the iceberg. Curr. Opin. Genet. Dev. 18, 317-323. Ö zbudak, E. M., Tassy, O. and Pourquie, O. (2010). Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc. Natl. Acad. Sci. USA 107, 4224-4229. Pourquié , O. (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650-663. Schrö ter, C., Ares, S., Morelli, L. G., Isakova, A., Hens, K., Soroldoni, D., Gajewski, M., Jü licher, F., Maerkl, S. J., Deplancke, B. et al. (2012). Topology and dynamics of the zebrafish segmentation clock core circuit. PLoS Biol. 10, e1001364. Shankaran, S. S., Sieger, D., Schrö ter, C., Czepe, C., Pauly, M.-C., Laplante, M. A., Becker, T. S., Oates, A. C. and Gajewski, M. (2007). Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and her15 reveal novel modes of expression and contribute to the segmentation clock. Dev. Biol. 304, 615-632. Takashima, Y., Ohtsuka, T., Gonzalez, A., Miyachi, H. and Kageyama, R. (2011). Intronic delay is essential for oscillatory expression in the segmentation clock. Proc. Natl. Acad. Sci. USA 108, 3300-3305. Tataroğ lu, O. and Schafmeier, T. (2010). Of switches and hourglasses: regulation of subcellular traffic in circadian clocks by phosphorylation. EMBO Rep. 11, 927-935. Trofka, A., Schwendinger-Schreck, J., Brend, T., Pontius, W., Emonet, T. and Holley, S. A. (2012). The Her7 node modulates the network topology of the zebrafish segmentation clock via sequestration of the Hes6 hub. Development 139, 940-947. Uriu, K., Morishita, Y. and Iwasa, Y. (2009). Traveling wave formation in vertebrate segmentation. J. Theor. Biol. 257, 385-396. DEVELOPMENT RESEARCH ARTICLE 4167