* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 24: the genetic code
Bottromycin wikipedia , lookup
Endomembrane system wikipedia , lookup
Gene regulatory network wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Western blot wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding RNA wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein adsorption wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene expression wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Epitranscriptome wikipedia , lookup
Amino acid synthesis wikipedia , lookup
List of types of proteins wikipedia , lookup
Biochemistry wikipedia , lookup
Transfer RNA wikipedia , lookup
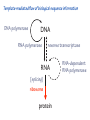
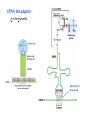
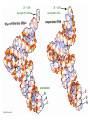
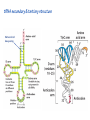
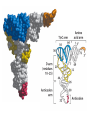
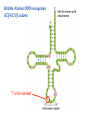
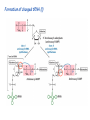
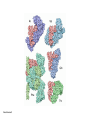
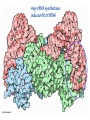
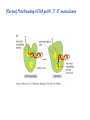
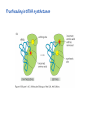
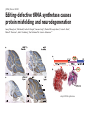
Lecture 24: the genetic code Key learning goals: • Know that 90% of a cell’s energy can be expended making • • • • • • • • • • • proteins Understand how to read the genetic code table. Be prepared to interconvert between DNA, mRNA, and polypeptide sequences. Understand the roles of mRNA, tRNA, and aaRS enzymes. Be able to explain the Brenner-Crick “adaptor hypothesis.” Be prepared to explain the secondary and tertiary structures of tRNA. Know how the anticodon works and what the 3’ stem does. Understand how the genetic code is embedded in aaRS enzymes (this is a critical point). Understand the “wobble” hypothesis. Understand how wobble works at the level of base-pairing. Be able to explain why adenosine deaminase enzymes are essential for the synthsis of wobble tRNA’s. Understand the biochemistry of aaRS enzymes. Understand how some aaRS’s proofread, and why. Template-mediated flow of biological sequence information DNA polymerase (2% RNA polymerase reverse transcriptase 62% [splicing] ribosome TVSXIMR RNA-dependent RNA polymerase Protein synthesis is a big deal. Proteins are the most abundant macromolecules in the cell. The vast majority of enzymes are proteins. Up to 90% of a cell’s energy can be directed toward protein synthesis. 25% of an E. coli cell’s dry mass can be ribosomes: almost 1,000 new ribosomes made per E. coli cell per minute! Many important antibiotics target protein synthesis. David Goodsell Transcription and translation facilitate regulation and amplification in gene expression One copy of the Ovalbumin gene many Ovalbumin mRNA’s 30,000,000,000,000,000,000 molecules of ovalbumin Definition: The GENETIC CODE is the set of RULES by which a linear sequence of nucleotides specifies the linear sequence of a polypeptide. (2% 62% But what is the nature of the genetic code? TVSXIMR y one reating nitrous s are amma), the ons in e code ature), ssible. obins hanges. le out must be correct ) along obvious omma’. ‘sense’, a-free about half the mutants made with base analogues are leaky) ; (2) Streisinger lo has found that whereas mutants of the lysozyme of phage T4 produced by How I Isaid totoyou, when you lysozyme baas-analogues usually leaky, How often often have haveare said you,that that whenall youhave have mutants produced by proflavin are negative, eliminated the impossible, whatever remains, that is, impossible, whatever remains, the however function improbable, is completely lacking. must be the truth. improbable, must be the truth. If an acridine mutant i,3 produced by, say, adding a —Sherlock Holmes base, it should revert to ‘lvild-type’—Sherlock by deleting a bass. Our work on revertants of FC-0 shows that it-usually Starlinq point 3 ,, ;$I Overlappirq +7 NUCLEIC ACID * I’ ’ ,-J+-~---- ’ code --’ ’ ’ ’ ETC. 1 3 Non-overlapplnq ' Code Fig. 1. To show the difference between an overlapping code and a non-overlappinu code. The short wrticnl lines represent the bases of the nucleic acid. The czw illustrated is for a triplet code Crick et al., (1961) Nature 192:1227 Reading along a DNA or RNA strand in one direction, there are THREE possible READING FRAMES: start +1 +2 CATTAGGAGACTCATTAGGAGACT CATTAGGAGACTCATTAGGAGACT CATTAGGAGACTCATTAGGAGACT Notes: • In double-stranded DNA there are SIX possible reading frames: three on the top strand, and three on the boom strand. • A +2 change in reading frame is equivalent to moving the frame -1. Which codons specify which amino acids? We understand the syntax, now we need to learn the vocabulary Decoding the genetic code Codon assignments You need to understand this table, but you don’t need to memorize it. stop. Start You need to understand this table, but you don’t need to memorize it. (2% 62% How is the genetic code implemented? TVSXIMR Medical Research Council Unit for the Study of the Molecular Structure of Biological Systerns, Cavendish Laboratory, Cambridge, England. ON DEGENERATETEMPLJATESAND THE ADAPTOR HYPOTHESIS F.H.C. Crick, Medical Research Council Unit for the Study of the Molecular Structure of Biological Systerns, Cavendish A Note for Laboratory, Cambridge, England. the RNA Tie Club. "1s there anyone so utterly lost as he that seeks a way where there is no way." 1955, unpublished (!) Kai Kg'us ibn Iskandar. Crick: adaptor hypothesis “…each amino acid would combine chemically, at a special enzyme, with a small molecule which, having a specific hydrogen-bonding surface, would combine specifically with the nucleic acid template. …there would be 20 kinds of adaptor molecule, one for each amino acid, and 20 different enzymes to join the amino acid to their adaptors. Sydney Brenner …calls this the “adaptor hypothesis”, since each amino acid is fitted with an adaptor to go to the template. tRNA: the adaptor (t is for transfer) Watson-Crick base pairing 3´ –OH acceptor site 3´ –OH acceptor site anticodon David Goodsell tRNA secondary & tertiary structure Watson-Crick base pairing RNA secondary structure: double helices similar to A-DNA A-DNA B-DNA tRNA David Goodsell RNA secondary structure: double helices similar to A-DNA B-DNA A-DNA Wobble Wobble: Alanine tRNA recognizes GC[A/C/U] codons: “I” is for inosine! Wobble How inosine promotes wobble pairing I = inosine hypoxanthine inosine base: hypoxanthine hypoxanthine Notice: inosine is deaminated adenosine! hypoxanthine tRNA synthetases assign the codons …there would be 20 kinds of adaptor molecule, one for each amino acid, and 20 different enzymes to join the amino acid to their adaptors. — F. Crick David Goodsell Do not memorize this table (unless you are very bored). Activation of amino acid %84 Formation of charged tRNA EQMRSEGMH X62%W]RXLIXEWI 44M EHIR]PEXIHEQMRSEGMH T]VSTLSWTLEXEWI 4MLIEX X62% X62%W]RXLIXEWI %14 EQMRSEG]PX62% Activation of amino acid mixed anhydride Formation of charged tRNA (I) Formation of charged tRNA (II) Activation of amino acid %84 Formation of charged tRNA EQMRSEGMH X62%W]RXLIXEWI 44M EHIR]PEXIHEQMRSEGMH T]VSTLSWTLEXEWI 4MLIEX X62% X62%W]RXLIXEWI %14 EQMRSEG]PX62% Three sources of error in translation: 1. aaRS uses wrong amino acid as substrate + 2. aaRS selects wrong tRNA as substrate + 3. Ribosome selects wrong aatRNA for codon Distribution of bases required for correct charging of tRNAs The diameter of each yellow circle indicates the number of different tRNA molecules for which that position is an aaRS specificity determinant. David Goodsell Asp-tRNA synthetase: induced-fit of tRNA David Goodsell Example: why it’s hard to acheive specificity in an aaRS Free energy (Δ∆G°´) of methylene binding is only ~12 kJ/mol Thus, @ equilibrium the synthetic site of Ile RS should contain ≥1 Val for every 200 Ile Example: why it’s hard to acheive specificity in an aaRS But: measured misincorporation by Ile RS is <1 Val per 3000 Ile, not >1 per 200. Something is increasing accuracy (reducing error) by >10-fold. (Review) Proofreading in DNA pol/III: 3´-5´ exonuclease Proofreading in tRNA synthetases Proofreading by aaRS’s - example %84 -PI -PI67 44M X62% -PIEHIR]PEXI -PI67 %14 %84 EQMRSEG]PX62% :EP -PI67 ATP and time are consumed in a “futile cycle” to increase accuracy! 44M :EPEHIR]PEXI X62% -PI67 %14 :EP X62% Proofreading by aaRS’s %84 Some aaRS’s proofread the aminoacyl-adenylate EQMRSEGMH X62%W]RXLIXEWI 44M EHIR]PEXIHEQMRSEGMH T]VSTLSWTLEXEWI 4MLIEX X62% X62%W]RXLIXEWI %14 EQMRSEG]PX62% Others proofread the aa-tRNA …and some don’t bother to proofread at all. ARTICLES sti cell mutation was mapped to chromosome 8 (s (Aars) gene. a, Thein amino-acid activation and tRNA aminoacylation, and an evoluacids, no differences viability were observed following cM ^ s.e.m.). b, The RP24-359N5 bacterial artificial chromosome tionarily conserved editing domain for hydrolytic editing of nonaddition of alanine, histidine or methionine (P . 0.05). In contr contains the Aars, Mrcl and Pdpr genes, of which the latter two wer cognate amino acids (glycine or serine) from misaminoacylated serine dramatically increased cell death of sti/sti fibroblasts in a do by partial digestion with NotI. Positions of primers used to genoty Ala (2006) Nature 443:50 (refs 10, 11). Analysis of the primary structure of AlaRS tRNA dependent manner relative to that observed in wild-type cultu suggested that Ala 734 lies within the putative editing domain (P , 0.0001). Mutant fibroblasts were marginally sensitive to glyc (Fig. 3a). The area encompassing this residue (Fig. 3a, b (shown in (2.59 ^ 1.22% versus 5.30 ^ 1.19% cell death in wild-type ver affected challenged serine than glycine. Thus, purple)) has been shown to enhance aminoacylation efficiency mutant cells;when values are meanwith ^ s.e.m.). Thewith specific sensitivity 11 examined the effect mutation had on of the en but has no role in amino-acid activation in Escherichia coli . The serine is consistent withthis defective editing inthe theability sti mutant Al Ala . With the mutant enzyme,todea hydrolyse human Ser-tRNAshow sti Ala–Glu substitution seems to be within a loop region well outside protein. Also, sti/þ fibroblasts an increased sensitivity ser Ala was diminished by a reproducible 40–50% (F of Ser-tRNA of the active site of the editing domain (.15 Å) but, nevertheless, that is intermediate to that 1of wild-type Ala and sti/sti fibrobl 1 Jeong Woong Lee1, Kirk Beebe2, Leslie A. Nangle2, Jaeseon Jang1†, Chantal M. Longo-Guess , Susan A. Cook , aboveofbackgrou contrast, no deacylation Ala-tRNA within the sequence predicted to form this domain (Fig. 3b). (Fig.In3h). This contrasts with theofnormal phenotype sti/þ m Muriel T. Davisson1, John P. Sundberg1, Paul Schimmel2 & Susan L. Ackerman1,3observed with either mutant or wild-type enzyme, ex In E. coli, mutations within the AlaRS editing domain result suggesting that environmental challenges can result in gene dos Ala Ala theofpossibility of a some loss of in editing specificity by the sti Ser-tRNA inMisfolded increased production of misacylated Gly-tRNA effects the sti mutation. proteins are associated with several pathological or conditions including neurodegeneration. Although (Supplementary 8). these abnormally folded proteins result from mutations in genes encoding disease-associated proteins (forFig. Defects in editing ofexample, non-cognate amino acids from mischar repeat-expansion diseases), more general mechanisms that lead to misfolded proteins in neurons remain largely A loss of editing activity should the acids producti tRNAs should lead to misincorporation ofresult these in amino dur unknown. Here we demonstrate that low levels of mischarged transfer RNAs (tRNAs) can lead to an intracellular release of misacylated tRNAs. In vivo, these incorrect prod protein synthesis, which would in turn lead to the accumulation accumulation of misfolded proteins in neurons. These accumulations are accompanied by upregulation of cytoplasmic proteins. To determine if serine-induced protein chaperones and by induction of the unfolded protein response. We report misfolded/unfolded that the mouse sticky mutation, which death in sti/sti cells is correlated with the accumulation of unfol causes cerebellar Purkinje cell loss and ataxia, is a missense mutation in the editing domain of the alanyl-tRNA synthetase gene that compromises the proofreading activity of this enzyme duringproteins, aminoacylation of tRNAs.ubiquitination These we analysed of proteins in serine-trea findings demonstrate that disruption of translational fidelity in terminally differentiated neurons leads to the fibroblasts pretreated with mitomycin C to inhibit cell division accumulation of misfolded proteins and cell death, and provide a novel mechanismdilution underlying ofneurodegeneration. misfolded proteins (Fig. 3i). In the absence of ser addition, polyubiquitinated proteins were greatly increased in st The genetic code is established in the aminoacylation reactions of particularly in the rostral cerebellum.indicating This degeneration is slowly fibroblasts, that misfolded/unfolded proteins accumu aminoacyl-tRNA synthetases, where each amino acid is linked to its progressive so that most Purkinje cells degenerate over the course of a in these cells. Ubiquitinated proteins increased in both mutant cognate tRNA that bears the anticodon triplet of the code. The rate of year, excluding the majority of these neurons in the caudally located misincorporation of amino acids into proteins is very low (estimated lobule X. wild-type cells on addition of serine to the media, indicating t at one error in every 103–104 codons)1,2, and this high accuracy To investigate the high nature concentrations of Purkinje cell loss, we performed of serine cause protein misfolding even in w results largely from the precision of aminoacylation reactions. In immunohistochemical studies with the apoptotic markers cleaved type cells, perhaps by interfering with AlaRS editing. Consistent w addition to tRNA recognition, aminoacyl-tRNA synthetases must caspase 3 and cleaved poly(ADP-ribose) polymerase (PARP). Purkinje discriminate between amino acids in the cellular pool. Generally, cells that were positivemisfolded for these markers were elevation, observed in mutant, protein levels of stress-inducible, cytosolic h amino acids with side chains that are bulkier than those of the but not wild-type, animals at four weeks of age (Fig. 1h–j and data shock protein 72 (HSP72) were increased in sti/sti fibroblasts, cognate amino acids are sterically excluded from the active sites of not shown). Further confirmation that these cells were undergoing on the addition tRNA synthetases, but smaller amino acids can fit into the active site apoptosis was obtainedalso fromincreased TUNEL (TdT-mediated dUTP nickof endserine to wild-type cells (Fig. 3 Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration pocket and be misactivated and mischarged. These misactivated adenylates or mischarged tRNAs are normally cleared by the editing function of aminoacyl-tRNA synthetases, encoded by a domain that is distinct from the domain for aminoacylation. If they are not cleared, genetic code ambiguity is introduced (that is, a given codon in the messenger RNA will specify incorporation of more than one amino acid, resulting in the production of ‘statistical polypeptides’)3–6. Here we report that an editing defect in a single tRNA synthetase in the mouse results in neurodegeneration associated with protein characteristics consistent with heterogeneous polypeptide production (Supplementary Fig. 1). These results provide a novel mechanism for the generation of misfolded proteins, which are labelling)-positive mutant Purkinje cells (Fig. 1k–m). The sti mutation disrupts AlaRS editing The sti mutation disrupts the Aars gene alanyl tRNA synthetase For functional analysis, weon introduced the A734E mutation into b A genome scan using polymorphic microsatellite markers affected the sti mutation to chromosome 8. F2 animals initially localized the mouse and the human enzyme, which is 92% identical (includ Further fine mapping demonstrated that sti resided in a 1.54Ala 734) to mouse2,932 AlaRS. Far-ultraviolet circular dichroism (farcentimorgan (cM) region (45 recombinants, meioses; Fig. Figure 3 | Mutant fibroblasts are2aselectively sensitive to serine. a, CD) spectral analysis purified wild-typestructures and mutant proteins and Supplementary Fig. 4).domains Expression(a) analysis ofof genes in the sti and their three-dimensional (b). The amin critical region failed toidentical reveal any differences between wild-type and minima and indistinguishable far-UV CD spec activation domain (red) and editing mutant cerebellar RNA. However, sequencing of34complementary DNA domain (purple) is modelled on suggesting that theandA734E substitution does not induce ma the synthetase Pyrococcus horikoshii free-standing AlaRS aeolicus AlaRS revealed a C-to-A nucleotide change in the alanyl-tRNA 23 changes local secondary structure of AlaRS , respectively. Blue spheres show(Supplement the location o domain gene (Aars) at nucleotide 2,201 of in thehomologue transcript in mutant mice—