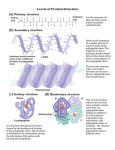
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Identification of a Chloroplast-encoded 9-kDa
Agarose gel electrophoresis wikipedia , lookup
Magnesium transporter wikipedia , lookup
Interactome wikipedia , lookup
Peptide synthesis wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression wikipedia , lookup
Size-exclusion chromatography wikipedia , lookup
Catalytic triad wikipedia , lookup
Gaseous signaling molecules wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Chloroplast DNA wikipedia , lookup
Protein purification wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Sulfur cycle wikipedia , lookup
Genetic code wikipedia , lookup
Biochemistry wikipedia , lookup
Western blot wikipedia , lookup
Biosynthesis wikipedia , lookup
Vol. 262, No. 26, Issue of September 15, pp. 12676-12684,1987 Printed in U.S.A. THEJOURNAL OF BIOLOGICAL CHEMISTRY 0 1987 by The American Society for Biochemistry and Molecular Biology, Inc. Identification of a Chloroplast-encoded9-kDa Polypeptide as a 2[4Fe-4S] Protein Carrying CentersA and B of Photosystem I* (Received for publication, April 13, 1987) Peter Bordier HejS, Ib Svendsed, HenrikVibe SchellerS, and Birger Lindberg MellerS From the $Department of Plant Physiology, Royal Veterinary and Agricultural University, 40 Thorvaldsemvej, DK-1871 Frederiksberg C and the §Department of Chemistry, Carlsberg Laboratory, 10 Gamk Carlsberg Vej, DK-2500 Valby, Denmark An improved procedure is reported for large-scale preparation of photosystem I (PS-I)vesicles from thylakoid membranes of barley (Hordeum uulgare L.). The PS-I vesicles contain polypeptides of molecular masses 82, 18, 16, 14, and9 kDa inan apparentmolar ratio of 4:2:2:1:2. The 18-, 16-, and 9-kDa polypeptides were purified to homogeneity after exposure of the PS-I vesicles to chaotropic agents. The isolated 9kDa polypeptide binds 65-70% of the zero-valence sulfur of denatured PS-I vesicles, and the remaining 30-3570 is bound to P700-chlorophyll a-protein1.The N-terminal amino acid sequence (29 residues) of the 9kDa polypeptide was determined. Comparison with the nucleotide sequence of the chloroplast genome of Marchantiapolyrnorpha (Ohyama, K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano, T., Sano, S., Umesono, K., Shiki, Y., Takeuchi, M., Chang, Z., Aota, S.-i., Inokuchi, H., and Ozeki, H. (1986) Nature 322, 572-574) and of Nicotiana tabucum (Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., Zaita, W., Chunwongse, J., Obokata, J., Yamaguchi-Shinozaki, K., Ohto, C., Torazawa, K., Meng, B. Y., Sugita, M., Deno, H., Kamogashira, T., Yamada, K., Kusuda, J., Takaiwa, F., Kato, A., Tohdoh, N., Shimada, H., andSugiura, M. (1986) EMBO J. 5, 2043-2049) identified the chloroplast gene encoding the 9-kDa polypeptide. We designate this gene psaC. The complete amino acid sequence deduced from the psaC gene identifies the 9-kDa PS-I polypeptide as a 2[4Fe-4S] protein. Since P700-chlorophyll a-protein1 carries center X, the 9-kDapolypeptide carries centers A and B. A hydropathy plot permits specific identification of the cysteine residues which coordinate centers A and B, respectively. Except for the loss of the N-terminal methionine residue, the primary translation product of the psaC gene is not proteolytically processed. P700-chlorophyll a-protein1binds 4 ironatoms and 4 molecules of acid-labile sulfide/molecule of P700. Each of the two apoproteins of P700-chlorophyll aprotein 1 contains thesequence Phe-Pro-Cys-Asp-GlyPro-Gly-Arg-Gly-Gly-Thr-Cys(Fish, L. E., Kuck, U., and Bogorad, L. (1985) J. Biol. Chem. 260, 14131421). The stoichiometry of the component polypeptides of PS-I indicates the presence of four copies of this sequence per molecule of P700. Center X may be * This research was supported in part by grants from the Danish Agricultural Research Council, the Danish Natural Science Research Council, Dansk Investeringsfond, the Thomas B. Thriges Foundation, the Carlsberg Foundation, the Tuborg Foundation, and Stiftelsen Hofmansgave and by a Niels Bohr grant from the Royal Danish Academy of Sciences and Letters. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solelyto indicate this fact. composed of two [2Fe-2S] centersbound to the 8 cysteine residues contained in these foursegments. The photochemical transferof electrons fromreduced plastocyanin to ferredoxin iscatalyzed by PS-I.’ The complex is known to contain P700, the reaction center chlorophyll of PS-I, and several electron acceptors which become reduced upon illumination. The signals obtainedby a large number of spectrophotophysicaltechniques (e.g. ESR and chemically induced dynamicelectronpolarization) haverevealed the involvement of at least five different electron acceptors denoted A,, AI, X, B, and A (1-3). Whereas spectrophotophysical techniques have been very useful in detecting these acceptors in complex preparations, the same techniques are of limited value in determining the identity of the chemical structures giving rise to the signals detected.As a result, the chemical identity of P700 and centers A,, and AI still remains unresolved (1-3). Mossbauer and ESR spectrometry indicated that centers B and A are iron-sulfur centers (4-7). Based on microwave power saturation studies, these two centers were further assigned as [4Fe-4S] clusters (8).The ESR spectrum of center X also had someresemblance to thoseof iron-sulfur centers (9), and Mossbauer spectroscopy suggestedthat center X could be a [4Fe-4S] center (10).However, microwave power saturation studies indicated that centerX was not a typical [4Fe-4S] or [2Fe-2S] center and that the spectrum possibly represented a chlorophyll anion magnetically interacting with iron (8). An alternative approach to study PS-I is to characterize the structural and functional role of each of the PS-Ipolypeptides. Using this strategy, H0j and Merller (11)and Golbeck (2) provided biochemical evidencedemonstrating that the82kDa polypeptides of P700-chlorophyll a-protein 1 bind an iron-sulfur center, most likely center X. PS-I preparations contain additional polypeptides of lower molecular mass (2, 3,11). Although the number reportedvaries, four polypeptides of approximate molecular masses 18,16,14, and9 kDa appear to belong to the PS-I core (2,3, 11).The 18-kDapolypeptide has beenclaimed to carry the iron-sulfur centers A and B (12, 13). However, this identification was based solely on correlation between the gradualdepletion of the 18-kDapolypeptide and the disappearance of centers A and B as monitored by ESR spectroscopy. Malkin et al. (14) isolated an 8-kDa polypeptide from chloroplasts of spinach (Spinacia oleracea). The The abbreviations used are: PS-I, photosystem I; DTT, 1,4dithiothreitol; Hepes, 4-(2-hydroxyethyl)-l-piperazineethanesulfonic acid; Mes, 2-N-(morpholino)ethanesulfonicacid; ORF, open reading frame; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; Tricine, N-[2-hydroxy-l,l-bis(hydroxymethyl)ethyl]glycine. 12676 A 9-kDa Protein of Photosystem I Carries Centers A and B preparation showed an ESR spectrum similar to that of a ferrodoxin but different from those of centers, A, B, and X. It remains to be established whether the reported spectra could have been derived from an artificial complex of similar molecular mass and generated from cysteine, sulfide, and iron. As demonstrated by Hej and Meller (ll), the coincidental comigration of such artificially formed oligomeric complexes with the 18 and 16-kDa polypeptides was previously interpreted to indicate that these two polypeptides were binding iron-sulfur clusters (15, 16). Lagoutte et al. (17) used in uiuo YS labeling and W-carboxymethylation to demonstrate that a 9-kDa polypeptide is the most cysteine-rich component in PS-I preparations obtained from spinach and showed that this polypeptide contains approximately 8 cysteine residues. Similar results pointing toward the 9-kDa polypeptide as an iron-sulfur protein of PS-I have been obtained in labeling studies with Anacystis nidulans (18). In contrast to these two studies, Bonnerjea et al. (12) found that a 30-40% depletion of the 16-, 14-, and clO-kDa polypeptides from a spinach PSI preparation did not result in a corresponding decrease in the ESR signals of centers A and B. These contradicting results called for a more direct approach for the identification of the apoprotein(s) of centers A and B. In this study, we have used acid-labile sulfide and zero-valence sulfur as specific markers for intact and degraded iron-sulfur clusters, respectively. The 18-, 16-, and 9-kDa polypeptides were purified to homogeneity from PS-I vesicles of barley after exposure of the vesicles to chaotropic agents. From its ability to bind zerovalence sulfur and from amino acid sequencing data, the purified 9-kDa polypeptide is identified as a chloroplastencoded, 2[4Fe-4S] protein that binds centers A and B. MATERIALS Unless AND METHODS otherwise indicated, all procedures were carried out at 4 “C. Preparation of PS-I the 9-kDa polypeptide but 12677 devoid of P700-chlorophyll a-protein 1 were combined and dialyzed against 5000 ml of 15 mM ammonium acetate (pH 5.0) 0.05% (w/v) Triton X-100. After dialysis (conductivity, 1.4 millisiemens), the material was loaded onto a column (0.9 x 30 cm) of CM-Sepharose (Pharmacia Biotechnology AB) equilibrated in 25 mM ammonium acetate (pH 5.5), 0.05% (w/v) Triton X-100 and eluted with a linear gradient of 25 mM ammonium acetate (pH 5.5), 0.05% (w/v) Triton X-100 and 400 mM ammonium acetate (pH 8.0), 0.05% (w/v) Triton X-100 (2 X 120 ml) at a flow rate of 10 ml/ h. The pH of the two buffers were adjusted with HOAc and NH3, respectively. Eluted fractions were again assayed for their content of acid-labile sulfide and zero-valence sulfur (20) and for their polypep- tide content. Fractions rich in the 9-kDa polypeptide were combined, lyophilized, dissolved in a minimal volume of AcA buffer, and finally applied to a column (1.6 x 95 cm) of AcA 34 equilibrated in AcA buffer and eluted with the equilibration buffer at a flow rate of 2 cm/ h. Fractions judged by SDS-PAGE to contain pure 9-kDa polypeptide were combined, lyophilized, and stored at -80 “C. 18- and 16-kDa Polypeptides-PS-I vesicles (146 ml) obtained from the AcA column were concentrated to a volume of 10 ml, diluted with 120 ml of 20 mM imidazole HCI (pH 7.6), and concentrated to 17 ml by ultrafiltration. Solid urea (7 g) was added to the concentrated PSI vesicles (16 ml, 1.20 mg of chlorophyll/ml), and the material was applied (25 ml/h) to a column (0.9 x 46 cm) of Polvbuffer Exchanger 94-(Pharmacia Biotechnology AB) equilibrated in 20 mM imidazole (pH 7.6), 5 M urea. Washing of the column with 160 ml of equilibration buffer served to elute the major part of the 18-kDa polypeptide originally bound to PSI. The column was subsequently washed-with 75 ml of 25 mM imidazole HCl (oH 7.6) and then with 60 ml of 25 mM imidazole HCl (pH 7.6), 0.1% (w/v) Triton X-100,0.05% Empigen BB (Albright & Wilson Ltd., Marchon Works, Whitehaven, Cumbria. United Kinadorn) which eluted some of the 16-kDa DOIVpeptide in a homogenous form. P700-chlorophyll a-protein 1, the 9and 14-kDa polypeptides, and some of the 16-kDa polypeptides were eluted by applying ‘I-fold diluted Polybuffer 74/HCl (pH 4.5), 0.1% (w/v) Triton X-100 to the column. The fractions containing P700chlorophyll a-protein 1 were concentrated by ultrafiltration and treated with NaSCN as described above for the 9-kDa polypeptide. These manipulations allowed the isolation of a particle which contained only P700chlorophyll a-protein 1 and the 14- and 9-kDa polypeptides. Vesicles Chloroplasts of barley (Hordeurn v&are L.) were isolated and osmotically lysed as described in Ref. 19. The lamellar systems (200400 mg of chlorophyll) were resuspended in 20 mM Hepes (pH 6.3), 15 mM NaCl, 5 mhi MgCl, at 2.0 mg of chlorophyll/ml. Triton X-100 (25 mg of 100% Triton X-lOO/mg of chlorophyll) was added, and after stirring for 30 min in the dark, a P700-enriched supernatant was obtained by centrifugation at 48,000 X g for 30 min. The super- Polyacrylamide Gel Electrophoresis to a column (5 x 20 cm) of DEAE-Sepharose CL-6B (Pharmacia Biotechnology AB, Uppsala, Sweden) equilibrated in the same buffer. The column was washed with 1 column volume of equilibration buffer containing 60 mM NaCl. The PS-I vesicles were-eluted using the eauilibration buffer fortified with a 60-500 mM linear NaCl aradient Analytical SDS-PAGE was carried out in slab gels at 6 “C according to Fling and Gregerson (21) with a 5% stacking gel (1.5 cm) and an 8-25 or 18% resolving gel (18 cm), both containing 0.1% SDS. Preparative PAGE was carried out in the same system except that the SDS concentration was lowered to 0.033% and the length of the stacking and resolving gel was 1 and 6 cm, respectively. When stated, the cysteine residues of the PS-I vesicle and isolated polypeptides were labeled by S-carbamoylmethylation using iodo[l-“Clacetamide in the presence of 2-mercaptoethanol and 8 M urea (22) before analysis by SDS-PAGE and autoradiography. Apparent molecular masses were deduced from the electrophoretic mobility of the following standards: catalase, aldolase, bovine serum albumin, ovalbumin, and cytochrome c. Determination of the amount of Coomassie Bril- (2 liant Blue R-250 bound to individual natant was immediately frozen at -80 “C or diluted 5-fold with 20 mM Tricine (pH 7.5), 0.2% (w/v) Triton X-100 and applied directly x 2,000 ml) at a flow rate of 110 ml/h. The P700-containing fractions eluted at an approximate NaCl concentration of 150 mM and were combined and concentrated by ultrafiltration to 30 ml in an Amicon cell (Amicon Corp.) fitted with a PM-30 membrane. The concentrated PSI material was applied to a column (5 X 95 cm) of AcA 34 (LKB-Produkter AB, Bromma, Sweden) equilibrated in 25 mM Mes (pH 6.5), 250 mM NaCl, 0.1% (w/v) Triton X-100 (AcA buffer) and eluted at a flow rate of 3 cm/h (11). The vield of P700 was typically 300 nmol. When needed, the PSI vesicles were concentrated by ultrafiltration using a PM-10 membrane. Isolation of Polypeptides 9-/Da Polypeptide-Solid NaSCN (3.2 g) was added to a concentrated suspension of PSI vesicles (10 ml, 1.32 mg of chlorophyll/ml), followed by gentle shaking for 30 min in the dark. The suspension was diluted to 15 ml with AcA buffer and immediately loaded onto a column (2.6 x 95 cm) of AcA 34 equilibrated in AcA buffer and eluted at a flow rate of 3 cm/h. Fractions (5.5 ml) were analyzed for their content of acid-labile sulfide and zero-valence sulfur (20), and their polypeptide content was monitored by SDS-PAGE, followed by Coomassie Brilliant Blue R-250 staining. Fractions (70-85) enriched in destaining Ball (23). of the polyacrylamide Amino Acid polypeptides gels was Analysis carried after staining and out according to and Sequencing Samples were hydrolyzed with 6 N HCl at 110 “C in sealed, evacuated tubes for 24 h, and the hydrolysates were analyzed using a Durrum D500 amino acid analyzer. Half-cystine was determined after performic acid oxidation (24) or after S-carbamoylmethylation. SCarbamoylmethylation of isolated polypeptides was carried out by dialyzing the purified proteins against several hundredfold excess of 20 mM ammonium acetate (pH 6.3), 0.05% (w/v) Triton X-100, followed by lyophilization. The lyophilized protein samnle was dissolved in 7 M guanidinium chloride, 0.2 M Tris/HCl (pH 8.0), 5 mM EDTA, and the solution was thoroughly flushed with nitrogen. DTT was added to give a final concentration of 10 mM. After incubation at 40 “C for 2 h, iodoacetamide was added to the sample to give a final concentration of 200 mM. After incubation followed by 24 h at 4 “C, the sample was extensively 20 mM ammonium acetate (pH 6.3), 0.05% (w/v) lyophilized. at 25 “C for 2 h, dialyzed against Triton X-100 and 12678 A 9-kDa Protein of Photosystem Lyophilized polypeptide material was easily dissolved in 30% (v/ v) HOAc. Amino acid sequences were determined with both a Beckman 890C spinning cup sequenator (25) and an Applied Biosystems gas-phase sequenator Model 479A usingthe program provided by the company. Phenylthiohydantoins were identified by reverse-phase high-pressure liquid chromatographyas described by Svendsen et al. (26). C-terminal amino acid analysis (27) was carried out by carboxypeptidase Y (649 ng) digestion of the S-carbamoylmethylated 9-kDa polypeptide (13 nmol) in a reaction mixture (150 pl) containing 40 mM Mes (pH6.5). 0.5% SDS, and 14 nmol of norleucine as an internal standard. Aliquots withdrawn at different times were acidified (pH 2.2) to stop the enzymatic reaction and used directly for amino acid analysis. Acid-labile Sulfideand Zero-valence Sulfur Acid-labilesulfide and zero-valencesulfurweredetermined as reported by H0j and Mdller (20) using the methylene blue procedure with EtOAc extraction steps included to avoid interference from of chlorophyll. To secure accurate spectrophotometric determination methylene blue, the absorption spectrum(500-750 nm) of each sample was recorded using an Aminco DW-2c spectrophotometer with a typicalfull-scale setting of 0.05. The absorbance at 660 nmwas determined from the spectra. Barley ferredoxin was isolated essentially as described (28) and used as a reference. Additional Analytical Procedures Thylakoids *S-labeledinvivowere obtained as previously described (11). Chlorophyll was determined according to Arnon (29). P700 wasquantitated from its ferricyanide-oxidized minus ascorbatereduced spectrum using an Aminco DW-2c spectrophotometer and an extinction coefficientof 64 mM" cm" (30). I Carries Centers A and B rn n ~ 1 2 3 4 5 6 7 8 9 195- 105- 23- 9- - - CBB- Ag CBB 35S FIG. 1. Analysis of the polypeptide composition of purified PS-Ivesicles and isolated polypeptides by SDS-PAGE. Electrophoresis was carried out overnight at 6 'C using an 8-25% high RESULTS Tris gradient gel. Unless otherwise indicated,the gel was stained with Comparedtoour previous study (ll),an ion-exchange Coomassie Brilliant Blue R-250( C B B ) .Lane 1 , purified PS-I vesicles; chromatography step hasbeen introduced in the preparation lane 2, isolated 18-kDa polypeptide;lane 3, isolated 16-kDa polypepof the PS-I vesicles from barley. This allows processing of tide; lane 4, isolated 9-kDa polypeptideafter extensive handling; lane 5,isolated 9-kDa polypeptide;lane 6, thylakoids; lane 7,PS-I vesicles large amounts of starting material and eliminates contamidevoid of the 18- and 16-kDa polypeptidesas obtained after NaSCN nation with chloroplast coupling factor (Fig. 1, lane I ) . The treatment of PS-I vesicles originally treated with urea (the bands isolation procedure here reported has routinely been used to werevisualizedby alkalinesilver staining); lane 8, purified PS-I isolatePS-I vesicles fromthylakoidscontaining several vesicles obtained from plants "S-labeled in uivo; lane 9, autoradioghundred milligrams of chlorophyll. The yield of P700 is ap- raphy of sample in lane 8. proximately 1 nmol/mg of chlorophyll of the thylakoids. This corresponds to a yield of 30%. The PS-I preparation used in The Coomassie Brilliant Blue R-250bound to theindividual Ref. 11 had a chlorophyll to P700 ratio of 110, whereas the polypeptide bands of the PS-I preparation was eluted from ratiois 60in thepresentpreparation.This difference is the gels and quantified spectrophotometrically(23). Normalexplained by the prolonged exposure time to Triton X-100 ization based on their apparent molecular masses and a uniform binding of Coomassie Brilliant Blue R-250 gave the caused by inclusion of the ion-exchange step. five main components: InthePAGEsystems previouslyused, the major PS-I following stoichiometryforthe PSpolypeptides were assigned molecular masses of 70 (doublet), 2.01.2:1.2:0.5:1.1, indicating a stoichiometry in the native polypeptide I complex of 42:2:1:2 for the apoproteinsof P700-chlorophyll 18, 15, 10, and 8 kDa (11). In this study, the composition of the isolated PS-I vesicles was analyzed in the a-protein 1 and the la-, 16-, 14-, and 9-kDa polypeptides, high Tris gel system of Fling and Gregerson (21). Although respectively. devoid of urea, this system proved superior in focusing the The relative distribution of sulfur amino acids among the low molecular mass polypeptides of PS-I. Based on the elec- PS-I polypeptides was assessed by electrophoresis and autotrophoretic mobility of known standards in the high Tris radiography of a n 3sS-labeled PS-I preparation obtainedfrom system, thecalculated apparent molecular masses of the major barley seedlings grown in the presenceof ["S]sulfate (Fig. 1, PS-I polypeptides were 82 (doublet), 18, 16, 14, and 9 kDa, lanes 8 and 9). In agreement with earlier observations (11,17, respectively. Apparent molecular masses of 82 kDa for the 33), the 18- and 14-kDa polypeptides were found to incorpotwo apoproteins of P700-chlorophyll a-protein 1 are in close rate small amountsof "S label, whereas the 16-kDapolypepagreement with the molecular massespredicted from the tide was not labeled at all. P700-chlorophyll a-protein 1 and nucleotide sequence of their genes (31). The band at105 kDa the 9-kDapolypeptide were both strongly labeled. The superrepresents P700-chlorophyll a-protein 1 which has not been imposition of the labeled band at 9 kDa with that obtained converted into the apoprotein. The band 195 at kDa is prob- by Coomassie Brilliant Blue R-250 staining (Fig. 1, lanes 8 (17, 34, 35) inability to ably a dimer of P700-chlorophyll a-protein 1. A minor com- and 9) shows thatthereported ponent migrating just above the 9-kDa polypeptide was ob- visualize the 9-kDa polypeptide by Coomassie Brilliant Blue served. In some preparations, an additional minor componentR-250 staining was not due to an intrinsic property of the was observed inthe 23-kDaregion. This component is thoughtprotein, but merely reflected the poor characteristics of the to representresidual amounts of light-harvesting chlorophyll- SDS-PAGE systems earlierused. Specific assessment of the of the PS-I protein I (32) and was not detectable in most content of cysteine residues in the individual PS-I polypeppreparations. tides was achieved by14C-S-carbamoylmethylationof the PS- A 9-kDa Protein of Photosystem I Carries Centers A and B 0" 12679 - I preparation from spinach(17), but is in agreementwith the published nucleotide sequences of the genes encoding thetwo apoproteins (36). 195An isolated intact iron-sulfur protein can be detected by its property to release acid-labile sulfide, whereas a denatured 105iron-sulfur protein may retain acid-labile sulfide in the form 02-0 of zero-valence sulfur (37). Incubation of the PS-I preparation with 3.4 M NaSCN and subsequent gel filtration on AcA 34 allowed collection of fractions containingvarying amounts of Y the 18-, 16-, and 9-kDa polypeptides as monitored by SDS9 " PAGE and Coomassie Brilliant Blue R-250 staining of each ;gz individual fraction. The fractions also contained zero-valence 14-= sulfur, whereas no acid-labile sulfur was detectable. The elu9tion of zero-valence sulfur correlatedwith that of the cysteinerich 9-kDa polypeptide, but not with that of the 18- and 16kDa polypeptides. Fractions rich in zero-valence sulfur were B: 1 4 pattern ~ A: CBB stain combined and dialyzed. Subsequent chromatography on CMFIG.2. Polypeptide composition of purified PS-I vesicles Sepharose CL-GB followed by a final gel filtration step on as monitored by Coomassie Brilliant and isolated polypeptides Blue R-250 staining ( A ) and autoradiography after I4C-S- AcA 34 resulted in a homogeneous preparation of the 9-kDa carbamoylmethylation ( B ) .Electrophoresis was carried out over- polypeptide. From both columns, the elution of the 9-kDa night at 6 "C using an 8-25% highTris gradient gel. Lane I , thylakoids polypeptide and zero-valence sulfur coincided. The homoge(not S-carbamoylmethylated) + I4C-labeled molecular mass stan- neity of the isolated 9-kDa polypeptide was assessed by SDSdards; lane 2, "C-labeled molecular mass standards; lane 3, purified PAGE, followed by Coomassie Brilliant Blue R-250 staining PS-I vesicles; lane 4, isolated 18-kDa polypeptide; lane 5,isolated 16kDa polypeptide; lane 6, isolated 8-kDa polypeptide; lane 7, purified (Fig. 1, lane 5 ) and autoradiography after I4C-S-carbamoylmethylation (Fig. 2, lane 6).The zero-valence sulfur contained PS-I vesicles. in the isolated 9-kDa polypeptide was stable todialysis at pH 5 and tolyophilization and was therefore bound covalently to the polypeptide backbone, most likely as a trisulfide (37). The isolated polypeptide lacked chromophores absorbing around 420 nm. Such chromophores are presentin native ferredoxins (37). Extensive handling of the isolated polypeptide, e.g. by repeated lyophilizations, tended to generate tracesof a component of slightly faster electrophoreticmobility (Fig. 1, lane 1 2 3 4 5 6 7 x -" " - - 1 2 3 4 5 6 7 /I 4). The procedure developed to purify the 9-kDa polypeptide also provided homogeneous preparations of the 18- and 16kDa polypeptides. However, large amounts of these two poly0 5 10 1 20 0 5 10 15 20 pl P S I VESCLES pl PS I VESICLES peptides were more easilyobtained after treatmentof the PSFIG.3. Quantitation of the amount of zero-valence sulfur I preparation with6 M urea. When such an extract was applied associated with the homogeneous preparation of the 9-kDa to a column of Polybuffer Exchanger 94 equilibrated in 20 polypeptide as compared to the content of acid-labile sulfidemM imidazole HCl (pH7.6), 5 M urea, the 18-kDa polypeptide of t h e in nativePS-I particles containing an identical amount a did not bind to the column and could be collected in 9-kDa polypeptide. A, standard curve indicating a linear relationship between the amount of PS-I vesicles (0.27 mgof chlorophyll/ homogeneous form (Fig. 1, lane 2). Upon subsequent washing ml) assayed and the total amount of acid-labile sulfide and zero- of the column with 25 mM imidazole HCl (pH 7.6), 0.1% valence sulfur detected; B, standard curve indicating the linear rela- Triton X-100, 0.05% Empigen BB, a proportion of the 16tionship between the amount of PS-I vesicles assayed and theamount kDa polypeptide was released in a homogeneous form (Fig. 1, of Coomassie Brilliant Blue R-250 bound to the 9-kDa polypeptide of the PS-I vesicle after SDS-PAGE. A and B, aliquots of a homoge- lane 3 ) . A preparation containing P700-chlorophyll a-protein neous preparation of the 9-kDa polypeptide were subjected to analyses 1 and low molecular mass polypeptides was obtained by eluidentical to those described above. In one such experiment, the 9- tion with Polybuffer 74 (pH 4.5). Subsequent treatment of kDa polypeptide was found to bind Coomassie Brilliant Blue R-250 this preparation with NaSCN followed by gel filtration recorresponding to an absorption of 0.033 a t 595 nm and to contain sulted in apreparation containingP700-chlorophyll a-protein zero-valence sulfur producing an absorption of 0.0022 at 660 nm. As 1 and the 14- and 9-kDa polypeptides (Fig. 1, lane 7). A illustrated on A and B, this particular experiment demonstrated that the isolated 9-kDa polypeptide binds zero-valence sulfur correspond- specific association of acid-labile sulfideor zero-valence sulfur ing to 32% of the amount of acid-labile sulfide present in PS-I vesicles with the 18- or 16-kDa polypeptides was not observed under containing an identical amount of the 9-kDa polypeptide. any of the isolation procedures tested. Even when purified, the 16-kDa polypeptide was not reactive toward i ~ d o - l - [ ' ~ C ] acetamide (Fig. 2, lune 5), whereasa weak labeling was I preparationprior to electrophoresis (Fig. 2). The 9-kDa 18-kDapolypeptide (Fig. 2, lane 4 ) . A polypeptide was by far the mostlabeled component, with less obtainedwiththe condition for the successful application of the purification label inP700-chlorophyll a-protein1,littleinthe18-kDa homogeneous preparations polypeptide, and none in the16- and 14-kDa polypeptides. It procedures reported here to obtain should be noted that the denaturing conditions used for the of the 18-, 16-, and 9-kDapolypeptides of PS-I is theuse of a reductive alkylation of PS-I resulted in a partial loss of the highly purified preparation of PS-I vesicles as the starting material. 14-kDa polypeptidewhich seems to aggregatemoreeasily The amountof zero-valence sulfur bound to theisolated 9than the rest of the PS-I components (Fig. 2, lane 3 ) . The presence of I4C label inthe P700-chlorophyll a-protein 1 kDa polypeptide was quantitated with native PS-I as a stanpolypeptides is in contrast to the results obtained with a PS- dard (Fig. 3). Increasing amounts of PS-I were subjected to 32x ,t I I A 9-kDa Protein of Photosystem I Carries Centers A and B 12680 SDS-PAGE. After electrophoresis, the Coomassie Brilliant *m In Blue R-250 specifically bound to the 9-kDa polypeptide band of each gel lane was quantitated spectrophotometrically (Fig. 3B) (23). Identical amounts of the PS-I preparation were m assayed for acid-labile sulfide and zero-valence sulfur (Fig. 3A), and two standard curves were constructed. In an analogous manner, the Coomassie Brilliant Blue R-250 bound to 3 the purified 9-kDa polypeptide afterSDS-PAGEandthe correspondingcontent of zero-valance sulfur were deter-401s mined. The yield of zero-valence sulfur obtained by assaying the 9-kDa polypeptide from two different preparations was 32 and 37%of that obtained when assaying an amount of -0,010 native PS-I containing exactly the same amount of 9-kDa PSI. However, when the native PS-I vesicle was subjected to a -0,005 NaSCN treatmentanalogous to that used to isolate the 9-kDa PS-I, the recovery of acid-labile sulfide was less than 60% -0 even after incubation with DTT. It is therefore evident that the native 9-kDa polypeptide must bind a major part of the GEL SLCE NUMBER acid-labile sulfide of the native PS-I particle. FIG.5. Relative distribution of zero-valence sulfur between To determine therelative distribution of zero-valence sulfur the lowmolecular mass polypeptides. Electrophoresis was carried between P700-chlorophyll a-protein 1and the 9-kDa polypep- out for 5.5 h at 6 "C using an 18%high Tris gradient gel. After SDStide under identical experimental conditions, the purified PS- PAGE, the gel was cut into 5-mm segments which were analyzed for I vesicles were subjected to preparative SDS-PAGE. After their content of zero-valance sulfur (So,0).The polypeptide content of each segment was analyzed by re-electrophoresis, and the amount electrophoresis, the gel was cut into narrow horizontal seg- of Coomassie Brilliant Blue R-250 bound to each polypeptide was ments. The contentof acid-labile sulfide and of zero-valence quantitated. In this particular experiment, the recovery of the 9-kDa sulfur was determined after D T T incubation as described polypeptide is low. (20). Small,equally sized parts of each segment were used for re-electrophoresis to establish the polypeptide composition in peptides of P700-chlorophyll a-protein 1. The apoproteins of the segments and to spectrophotometrically quantify polythe P700-chlorophyll a-protein 1 barely enter the 18%resolving peptides present (23).When an 8-25% gradient gel was used, gel, which is therefore not suitable for determination of the between 30 and 35% of the recovered acid-labile sulfide was relative distribution of zero-valence sulfur betweenP700associatedwith P700-chlorophyll a-protein 1, whereas 65- chlorophyll a-protein 1 and the low molecular weight poly70% was associated with polypeptides in the low molecular peptides. mass region (Fig. 4). To separate the9-kDa polypeptide more It was of interest to obtain sequence information on the efficiently from those at 18, 16, and 14 kDa, a similar exper- isolated 9-kDa polypeptide. The S-carbamoylmethylated 9iment using an 18%resolving gel was performed (Fig. 5). This kDa polypeptide was therefore subjected to Edman degradaexperimentdemonstratedthatthe acid-labilesulfide re- tion in a liquid-phase spinning cup sequenator (-10 nmol, 29 covered after incubation with D T T was derived from the 9- cycles, repeated three times, 94% repetitive yield) as well as kDa polypeptide. Thus, after separation of the polypeptides in a gas-phase sequenator (-1 nmol, 92% repetitive yield). of the PS-Ivesicle by SDS-PAGE, 65-70% of the acid-labile The sequence for the 29 N-terminal residues of the isolated sulfide recovered after DTT treatment is associated with the 9-kDa polypeptide is shown on Fig. 6. The spacing of the 4 9-kDa polypeptide, whereas 30-35% resides in thelarge poly- identified cysteine residues is strongly indicative of a [4Fe4S] protein(38).Amino acidanalysis of the 9-kDapolypeptide I I after treatment with performic acid (24) revealed the presence 0,008 'BAD" of 8 cysteine residues/78 amino acids (Table I), indicating 8 2 a 16ADa that the protein mightbe a 2[4Fe-4S] protein. Very recently, 142Da the complete nucleotide sequences of chloroplast DNA from the liverwort Marchantia polymorpha (39) and from tobacco (Nicotiana tabacum) (40) have been determined. Ohyama e t al. (39) pointed out that the M. polymorpha sequence contained two ORFs denotedfrxA and frxB inwhich the periodic appearance of cysteine residues resembled that of [4Fe-4S] ferredoxins. When the N-terminal sequences predicted from these two ORFs were compared with the N-terminal amino acid sequence of the 9-kDa polypeptide isolated from PS-I vesicles of barley, the homology with frxA, but not with frxB, was strikingand leaves nodoubtthattheisolatedPS-I polypeptide is coded for by the corresponding ORF on the barley chloroplast genome (Fig. 6). To accord with the no0 menclature used by Ohyama et al. (39) and Gray e t al. (41) 1 3 5 7 0 1 1 1 3 GEL SLICE NUMBER for the previously identified chloroplast genes encoding memFIG. 4. Relative distribution of zero-valence sulfur between brane proteins catalyzing light reactions of photosynthesis, P700-chlorophylla-protein 1 and the low molecular mass we designate this gene as psaC. The psaCof the M. polymorpolypeptides. Electrophoresis was carried out for 5.5 h at 6 "C using an 8-25% high Tris gradient gel. The bars indicate the polypeptide pha chloroplast genome is locatedbetween the ndh41 and distribution between the different gel slices (5 mm) as monitored by ndh4 genes (39). An initial search on the chloroplastgenome of tobacco (40) for a similar ORF coding for an N-terminal re-electrophoresis. -i ; - l l i I i A 9-kDa Protein of Photosystgm I Carries Centers A and B FIG. 6. Partial amino acid sequence for the isolated 9-kDa 2[4Fe-4S] polypeptide of PS-I from barley compared with the amino acid sequences deduced from the corresponding gene psaC on the chloroplast genomes of tobacco (40) and M. polyrnorpha (39).Identity between codons and amino acid residues of the 9-kDa polypeptide from different species is indicated with a plus. Differences between the two nucleotide sequences causing amino acid substitutions are indicated by dashed bores. The cysteine residues chelating the [4Fe-4S] clusters are bored. The nucleotide inserted into the tobacco sequence (40) to restore identity with the N-terminal amino acid sequence of the 9-kDa polypeptide from barley is indicated with a question mark. TABLE I Amino acid composition of the 9-kDa photosystem Ipolypeptide carrying the two I4Fe-4SI centers A and B Amino acid H. vulgare N . tabacurn" M . polymorphab 9 8 9 Cys' 8.1 7 6 5 Asx 5.6 7 6 7 Thr 6.1 5 7 6 Ser 7.0 6 6 7 Glx 7.2 4 4 4 Pro 3.6 5 4 6 GlY 6.2 6 6 6 Ala 6.3 6 6 5 Val 4.8 3 3 1 Met 1.2 4 4 4 Ile 3.5 4 4 4 Leu 4.1 3 3 3 TYr 2.5 1 1 P he 1.8 2 1 His 1.3 1 2 4 3 4 LYS 4.0 6 5 5 Arg 4.7 2 1 ND Trp NDd 12681 of codon 20 in the psaC ORF of M. polymorpha restored a nucleotide sequence whichcoded for a protein identical to the N-terminal part of the barley 9-kDa protein (Fig. 6). The amino acid composition of the isolated 9-kDa PS-I polypeptide of barley resembles that predicted from the psaC genes between of M . polymorpha and tobacco (Table I). The identity the partial sequence of the 9-kDa PS-I of barley and the sequencededuced from the psaCgene of tobacco strongly indicates that the sequence of the remaining part of the barley protein will be very homologous to the correspondingsequences in tobacco and M . polymorpha. The C-terminal amino acid of the 9-kDa polypeptide was determined by carboxypeptidase Y digestion (27) and was found tobe tyrosine. Thus, apartfrom the removal of the Nterminal methionine residue, the primary translation product of the 9-kDa polypeptide is not proteolytically processed. Xray crystallographic analyses of the soluble 2[4Fe-4S] ferredoxin fromPeptococcus aerogenes had established the identity of the cysteines bound to the two [4Fe-4S] clusters (42). One [4Fe-4S] cluster is bound to Cys-X-X-Cys-X-X-Cys (where X represents amino acid)in the N-terminalhalf of the protein and to Cys-Pro in the C-terminalhalf. The second cluster is bound to Cys-X-X-Cys-X-X-Cys in the C-terminalhalf and to Cys-Pro in the N-terminal part of the protein. The partial amino acid sequence of the isolated 9-kDa polypeptide and the deduced amino acid sequence for the corresponding proteins in tobacco and M . polymorpha reveal identical cysteinecontaining segments, thereby identifying these proteins as 2[4Fe-4S] proteins. A hydropathy plot according to Hopp and Woods (43) is shown in Fig. 7. It is interesting to note that the 4 cysteine residues at positions 10, 13, 16, and 57 anchoring one of the [4Fe-4S] centers(42) are positioned in relatively hydrophobic regions, whereas the cysteineresidues at positions 20, 47, 50, and 53 coordinating the second center are located in more hydrophilic stretches of the molecule. The positioning of the cysteine residues in the bacterial 2[4Fe-4S] ferredoxins (38) suggests that the cysteine residue at position 33 is not involved in anchoring an iron-sulfur center. The aminoacid composition of the isolated 16- and 18-kDa r I l l I I I l l 1 Total 78.0 - 79 81 81 Deduced from the nucleotide sequence identified on the genome of N . tabacurn (40) after insertion of a missing nucleotide (see "Results"). * Deduced from the nucleotide sequence identified on the genome of M.p o l y m o r p h (39). Determined as cysteic acid after performic acid oxidation (24). ND, not determined. a part of the polypeptide homologous to thebarley 9-kDa polypeptide was unsuccessful. Search at the nucleotide level, however, revealed extensive homology between the psaCregion of M . polymorpha and the corresponding region in tobacco. It turned out that the ORF found in M . polymorpha was not listed in the analysis of the tobacco chloroplast genome, most probably because a single base pair had been missed in the sequencing of the tobacco chloroplast genome. Thus, insertion of a thymine residue in the noncoding strand of the tobacco sequence at a position corresponding to the wobble position 1 i0 20 40 30 50 60 AMINOACIDNUMBER 70 &J FIG. 7. Hydropathy plots of the 9-kDa 2[4Fe-4S] polypeptides of tobacco and M. polyrnorpha carrying centers A and B of photosystem I. The amino acid sequences werederived by identification of the gene psaC for the two polypeptides on the completely sequenced chloroplast DNA of tobacco (40) and M.polymorphs (39). The hydropathy plots were calculated according to Hopp and Woods (43). A 9-kDa Proteinof Photosystem I Carries CentersA and B 12682 SIWC. P. elrdenli P . elldenti Reriduer 11-281 iz9-541 FIG. 8. Comparison of the amino acid sequences of the 9kDa 2[4Fe-4S] PS-I polypeptides of barley and tobaccowith that of a 2[4Fe-4S] ferredoxin of P. ehdenii (38).To indicate the presumed gene duplication (47), the N-and C-terminal halves of the proteins have been aligned with respect to the positioning of the cysteine residues chelating the two [4Fe-4S] clusters. Identicalresidues are indicated with bores. Each amino acid is indicated by the standard single-letter code. polypeptides was also determined.' The 16-kDa polypeptide contained neither methionine nor cysteine, in accordance the earlier reported absenceof label in this protein afterI4Ccarbamoylmethylation and after labeling with 35Sin vivo (11). This polypeptide also lacks histidine. The 18-kDa polypeptide contained small but significant amountsof cysteine (-1 residue/molecule).2 Thus, the 16-and 18-kDa polypeptides are not iron-sulfur apoproteins. Both the 18- and 16-kDa polypeptides contained very high levels of alanine and proline. This was reflected in the partial aminoacid sequences which we have obtained for these two proteins.* DISCUSSION Partial amino acid sequencing of the 9-kDa polypeptide isolated from PS-I vesicles of barley permitted identification of its correspondinggene psaC on the chloroplast genomes of of the tobacco (40) andM . polymorph (39) and identification protein as a carrier of two [4Fe-4S] clusters (Fig. 6). The spacing of the cysteine residues does not fit that of soluble [ZFe-ZS] proteins which also lack the Cys-Pro segment (38, 44-46). The soluble2[4Fe-4S] ferredoxins reveala strong internal homology presumably due toa gene duplication (47). The deduced sequence for the 9-kDa polypeptide of tobacco displays a similar internal homology between residues 3-25 and 40-62 (Fig. 8). The availability of analytical procedures(20) permitting fast andreliable determination of acid-labile sulfide and zerovalence sulfur was essential in the development of the procedure which resulted in isolation of the 9-kDapolypeptide from barleyas a partiallydenatured 2[4Fe-4S] protein.Partial denaturation was evidenced by the lack of absorption around 420 nm andby the inabilityof the polypeptide to release acidlabile sulfide without prior reduction. The amount of zerovalence sulfur bound to the isolated 9-kDa polypeptide was quantified by two different procedures. After SDS-PAGE of the purified PS-I vesicle, the amount of zero-valence sulfur associated with the 9-kDapolypeptide was twice the amount found tobe associated withP700-chlorophyll a-protein 1 (Fig. 4). In its purified state, the amount of zero-valence sulfur associated with the 9-kDa polypeptide was one-third of the amount of acid-labile sulfide present in a native PS-I preparation containing an identical amount of 9-kDa polypeptide (Fig. 3). This is explainedby the less than 60% yield of acidlabilesulfide obtained from denaturedPS-I vesicles after D T T reduction. Incomplete conversion of zero-valence sulfur into acid-labile sulfide upon reduction may be one reason for the lower recovery of acid-labile sulfide from the isolated 9kDa polypeptide. Thus, the yield obtained with cysteine trisulfide as a standard was 77% (37). In addition, Petering et * H. V. Scheller, P. B. Hej, I. Svendsen, and B. L. Mller, manuscript in preparation. al. (37) observed that the zero-valence sulfurs bound in the oxidatively denatured bacterial 2[4Fe-4S] ferredoxins of Micrococcus lactylyticus, Clostridium pasteurianum, and Peptostreptococcus elsdenii were only 42, 48, and 63% of the acidlabile sulfide found in the native proteins, respectively. Of these soluble ferredoxins, that of P. elsdenii showsthe highest degree of structural homology with the psaC gene product (Fig. 8) (38). The low recoveries led Petering etal. to conclude that thezero-valence sulfur of these proteins is bound mainly in a cysteine trisulfide structure (37). Quantitative retainment of the acid-labile sulfide originally present in native [4Fe-4S] proteins would require the formationof a cysteine tetrasulfide structure (37). However, the recovery of zero-valence sulfur from oxidatively denatured [2Fe-2S] proteins wasalso low (37). In contrast to the results of Petering et al. (37) and to the Golbeck and Kok (48) have with results obtained in this study, reported a 100% recovery of acid-labile sulfide following denaturation of PS-I particles and regeneration of acid-labile sulfidefromzero-valence sulfur by D T T treatment. Using recoveries of 63 and 77% for the formation of zero-valence sulfur and the regeneration of acid-labile sulfide, respectively, the amount of acid-labile sulfur calculated to correspond to the amount of zero-valence sulfur detected on the isolated 9kDa PS-I polypeptide corresponds t o 70% of the acid-labile sulfide inthenativePS-Iparticle.This value is inclose agreement with the relative distribution of zero-valence sulfur polypepbetween P700-chlorophyll a-protein 1and the 9-kDa tide as determined after the denaturing conditions of SDSPAGE (Fig. 4) by which acid-labile sulfide is converted into zero-valence sulfur (11, 20, 34, 35). Native PS-I vesicles are generally foundtocontain 12 molecules of acid-labile sulfide for each molecule of P700 (1, 2, 3, 11).From the results presentedhere, we can assign 8 of the molecules of acid-labile sulfide to the 9-kDa polypeptide and the remaining 4 to the apoproteins of P700-chlorophyll a-protein 1. H0j and Maller (11)and Golbeck and Cornelius (49) have recentlydemonstratedthat P700-chlorophyll aprotein 1 carries center X. Centers A and B are therefore identified as the two [4Fe-4S]clusters of the chloroplastencoded 9-kDa 2[4Fe-4S] polypeptide. Experiments based on ESR spectrometry have revealed a differential sensitivity of centers A and B toward oxidative denaturation(48)and toward reactivity with mercurials (50), with center B as the most sensitive cluster. Similarly, center B has been demonpstrated tobe sensitive to the membrane-impermeant probe diazonium benzene sulfonate (51). A hydropathy plot of the 9-kDa polypeptide reveals that the [4Fe-4S] cluster coordinated by cysteine residues 10, 13, 16, and 57 is buried in the membrane, whereas the cluster coordinatedby cysteine residues 20,47,50, and53 is more external (Fig. 7). We therefore conclude that these two [4Fe-4S] clusters represent centersA and B, respectively.Selective destruction of one[4Fe-4S] center had also been reported in the soluble 2[4Fe-4S] ferredoxinI of Azotobacter uinelandii(52). The localization of centers A and B on the same polypeptide chain is in agreement with the strong interaction observed between these two centers by ESR spectroscopy ( 5 , 7, 54). One important aspect regarding the biosynthesis of the 2[4Fe-4S]holoprotein is theformation of theiron-sulfur cluster.Denatured soluble[2Fe-2S] ferredoxinsare easily reconstituted (55). Although N- and C-terminal analyses of the isolated 9-kDa polypeptide established that the primary gene product of psaC is not post-translationally cleaved except for the loss of the N-terminal methionine residue, it has not yet been possible by reconstitution experiments to regenerate the ESR signals of the iron-sulfur centers from denatured PS- A 9-kDa Protein Photosystem of I Carries Centers A and B 12683 peptides were also purified to homogeneity. The function of I vesicles (56). Takahashi et al. (57) have recently demonstrated that sulfur atomsof the iron-sulfur clusterof chloro- these two polypeptides remains unknown. However, partial plast ferredoxin are derived from cysteine and thata soluble amino acid sequencing indicated that the two polypeptides stroma enzyme isinvolved in the cluster formation. It will be are related.* interesting to test the activity of this enzyme toward the isolated 9-kDa apoprotein. Acknowledgments-Hanne Linde Nielsen, Inga Olsen, Bodil Corneliussen, Lone Sbrensen, and Pia Breddam are thanked for skillful P700-chlorophyll a-protein 1 was showninthepresent technical assistance. Drs. Birte Svensson and David Simpson are study to carry 4 of the 12 molecules of acid-labilesulfide for helpful discussions. Professor Knud W. Henningsen and associated with the native PS-Ivesicle per molecule of P700. thanked Drs. Jan Lembeck, T. G . Petersen, and C.-E. Olsen are thanked for We have previously reported that P700-chlorophyll a-protein performing the computer analyses. 1binds 4.3 iron atoms/molecule of P700 (11). Using experimentalconditions whereP700-chlorophyll a-protein 1 had REFERENCES been functionally detached from the lower molecular mass 1. Rutherford, A. W., and Heathcote, P. (1985) Photosynth. Res. 6 , polypeptides, Golbeck and Cornelius (49) obtained absorbance 295-316 transients at 698 nm, indicating that the iron-sulfur center 2. Golbeck, J. H. (1987) J. Membr. Sci., in press associated with P700-chlorophyll a-protein 1 was center X. 3. Meller, B. L., H@j,P. B., Halkier, B. A., Olsen, I., Nielsen, H. L., and Madsen, A. (1987) Dan. J. Agron. 132,5-21 Mossbauer spectroscopyidentified center X as a[4Fe-4S] 4. Evans, E. H., Rush, J. D., Johnson, C. E., and Evans, M. C. W. center (10). However, extended x-ray absorption fine-struc(1979) Biochem. J. 182,861-865 ture measurements (58) and core extrusion studies (59) indi5. Malkin, R., and Bearden, A. J. (1971) Proc. Natl. Acad. Sci. cated that PS-I contains [2Fe-2S] clusterswell as as [4Fe-4S] U. S. A. 68, 16-19 clusters. In this study, centers A and B have been identified 6. Evans, E. H., Cammack, R., and Evans, M. C.W. (1976) Biochem. as [4Fe-4S] centers. This points toward X center as a [2Fe-2S] Biophys. Res. Commun. 6 8 , 1212-1218 7. Evans, M. C. W., Telfer, A., and Lord, A.V. (1972) Biochim. center. With 4 iron and 4 sulfuratomspresentonP700Biophys. Acta 2 6 7 , 530-537 chlorophyll a-protein 1 per molecule P700, this would permit 8. Rupp, H., Rao, K. K., Hall, D. O., and Cammack, R. (1978) the location of two [2Fe-2S] centers on this protein. Recently, Biochim. Biophys. Acta 5 3 7 , 255-269 Bonnerjea and Evans (60) have provided evidence for a cor9. Evans, M. C. W., Sihra, C. K., and Cammack, R. (1976) Biochem. responding heterogeneityof the signal associated with center J. 1 5 8 , 71-77 10. Evans, E. H. Dickson, D. P. E., Johnson, C. E., Rush, J. D., and X. Evans, M. C. W. (1981) Eur. J . Biochem. l l 8 , 8 1 - 8 4 If center X is composed of two traditional [2Fe-2S] centers, this would require the availability of 8 cysteine residues. P700- 11. H0j, P. B., and Mbller, B. L. (1986) J. Biol. Chem. 2 6 1 , 1429214300 chlorophyll a-protein 1 is composed of two apoproteins with 12. Bonnerjea, J., Ortiz, W., and Malkin, R. (1985) Arch. Biochem. approximate molecular masses of 83 kDaand which are Biophy~.2 4 0 , 15-20 (31). Both apoproteins present in near equimolar amounts 13. Golbeck, J. H., Parrett, K. G., and Root, L. L. (1987) in Progress in Photosynthesis Research (Biggins, J., ed) Vol. I, pp. 253-256, arechloroplast-encoded,andtheir genes (psaA and psaB) Martinus Nijhoff, Dordrecht, The Netherlands have been sequenced inmaize (36), tobacco (40), spinach (61), 14. Malkin, R., Aparicio, P. J., and Arnon, D. I. (1974) Proc. Natl. pea (62), M . polymorpha (39), Euglena (63), and SynechococAcad. Sci. U. S. A. 7 1 , 2362-2366 cus (64). In all these species, both genes specifiedthe following 15. Hiller, R. G., Mdler, B. L., and H@yer-Hansen,G. (1980) Curlscompletely conserved stretch of 12 amino acids: Phe-Pro-Cysberg Res. Commun. 4 5 , 315-328 Asp-Gly-Pro-Gly-Arg-Gly-Gly-Thr-Cys. Besides the 2cys16. Mdler, B. L., H@yer-Hansen,G., and Hiller, R. G. (1981) Proc. Int. Congr. Photosynth. Res. 3,245-256 teines of this segment, no other cysteines were found in the psaB gene product. ThepsaA gene of tobacco, maize, and pea 17. Lagoutte, B., Setif, P., and Duranton, J. (1984) FEBS Lett. 1 7 4 , 24-29 specified 2 additional cysteines, whereas thatof Euglena and 18. Guikema, J., and Sherman, L. (1982) Biochim. Biophys. Acta Synechococcus specified only 1 of these residues. Fish et al. 681,440-450 (53) speculated that the 2 cysteine residues of the conserved 19. Mdler, B. L., and Hplj, P. B. (1983) Carkberg Res. Commun. 4 8 , 161-185 segment indicated above were connected by a disulfide bond in the native protein. We propose, in agreement with specu- 20. H0j, P. B., and Mdler,B. L. (1987) Anal. Biochem. 2 4 0 , in press lations of Golbeck et al. (59), that these conserved cysteine 21. Fling, S. P., and Gregerson, D. S. (1986) Anal. Biochem. 1 5 5 , 83-88 residuesfromtwopsaApolypeptides and fromtwo psaB 22. Crestfield, A. M., Moore, S., and Stein, W. H. (1963) J. Biol. polypeptides constitute the 8 cysteine residues necessary to Chem. 238,622-627 coordinate the two [2Fe-2S] clusters of P700-chlorophyll a- 23. Ball, E. H. (1986) Anal. Biochem. 155,23-27 protein 1. Thus, four large polypeptides would be required to 24. Hirs, C . H. W. (1967) Methods Enzymol. 1 1 , 197-199 bind 1 molecule of P700 and thetwo [2Fe-2S] clusters repre- 25. Johansen, J. T., Overballe-Petersen, C., Martin, B., Hasemann, V., and Svendsen, I. (1979) Carlsberg Res. Commun. 4 4 , 201senting center X. Assuming a uniform binding of Coomassie 217 Brilliant Blue R-250, the stoichiometryof the polypeptides of 26. Svendsen, I., Martin, B., and Jonassen, I. (1980) Carlsberg Res. the native PS-I vesicle was calculated t o be 42:2:1:2 for the Commun. 45,79-85 apoproteins of P700-chlorophyll a-protein 1 and the 18-, 16-, 27. Martin, B., Svendsen, I., and Ottesen, M. (1977) Carlsberg Res. Commun. 42,99-102 14-, and 9-kDapolypeptides, respectively. These ratios arein close agreement with those obtained with I4C-labeled prepa- 28. Buchanan, B. B., and Arnon, D. I. (1971) Methods Enzymol. 2 3 , 413-440 escaped our atten- 29. Arnon, D. I. (1949) Plant Physiol. 2 4 , 1-14 rations from Synechococcus (56). It has not tionthattheseratiospredictthe presence of two 9-kDa 30. Hiyama, T., and Ke, B. (1972) Biochim. Biophys. Acta 2 6 7 , 160171 polypeptides/molecule of P700, which would give a calculated content of20 eq of iron and acid-labile sulfide/molecule of 31. Fish, L. E., and Bogorad, L. (1986) J. Biol. Chem. 2 6 1 , 81348139 P700. The experimentally found ratio is 12 (1, 2, 3, 11). At 32. Lam, E., Ortiz, W., and Malkin, R. (1984) FEBS Lett. 1 6 8 , 10present, we are not able to discriminatebetween the possible 14 explanations for this discrepancy. 33. Nechushtai, R., andNelson, N. (1981)J.Bid. Chem. 256,11624During the course of this work, the 18- and 16-kDa poly11628 12684 A 9-kDa Protein of Photosystem I Carries Centers 34. Mdler, B. L., Halkier, B. A., and H#j, P. B. (1987) in Progress in PhotosynthesisResearch (Biggins, J., ed) Vol. 11, pp. 49-52, Martinus Nijhoff, Dordrecht, The Netherlands 35. H0j, P. B., Halkier, B. A., andMdler, B. L. (1987) in Progress in (Biggins, J., ed) V O ~ .11, pp. 53-56, ~ h o t o ~ y n t h e s Research is Martinus Nijhoff, Dordrecht, The Netherlands 36. Fish, L. E., Kuck, U., and Bogorad, L. (1985) J . Biol. Chern. 260, 1413-1421 37. petering, D.,F ~ J.~A.,, and palmer, G. (1971) J , ~ i ~then. l , 246,643-653 38. Yasunobu, K. T., and Tanaka, M. (1980) Methods Enzyrnol. 6 9 , 228-238 39. Ohyama, K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano,T., S a m S., Umesono, K., Shiki, Y., Takeuchi, M., Chaw, Aota, S.-i., Inokuchi, H., and Ozeki, H. (1986) Nature 3 2 2 , 572-574 40. Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., Zaita, N., Chunwongse, J., Obokata, J., Yamaguchi-Shinozaki, K., Ohto, C., Torazawa, K., Meng, B. Y.,Sugita, M., Deno, H., Kamogashira, T., Yamada, K., Kusuda, J., Takaiwa, F., Kato, A., Tohdoh, N., Shimada, H., and Sugiura, M. (1986) EMBO J. 5 , 2043-2049 41. Gray, J. C., Blyden, E. R., Eccles, C. J., Dunn, P. P. J., Hird, S. M., Hoglund, A.-S., Kaethner, T. M., Smith, A. G . , Willey, D. L., and Dyer, T. A. (1987) in Progress in Photosynthesis Research (Biggins, J., ed) Vol. IV, pp. 617-624, Martinus Nijhoff, Dordrecht, The Netherlands 42. Adman, E. T., Sicker, L. C., and Jensen, L. H. (1973) J . Bioi. Chem. 248,3987-3996 43, H ~ T. ~ p., and ~ Woods, , K, R. (1981) proc, Natl, ~ ~ sei. ~ IJ. S. A. 78, 3824-3828 44. Bruschi, M. H., Guerlesquin, F. A., Bovier-Lapierre, G. E., Bonicel, J. J., and Couchoud, P. M. (1985) J. Biol. Chern. 260, 8292-8296 45. Takahashi, Y., Hase, T., Wada, K., and Matsubara, H. (1983) Plant Cell Physiol. 2 4 , 189-198 46. Hase, T., Matsubara, H.,Koike, H., and Kat& s. (1983) Biochirn. Biophys. Acta 7 4 4 , 46-52 47. Tanaka, M., Nakashima, T., Benson, A., Mower,H., and Yasunobu, K. T. (1966) Biochemistry 5 , 1666-1681 z., A and B 48. Golbeck, J. H., and Kok, B. (1978) Arch. Biochern. Biophys. 188, 233-242 49. Golbeck, J. H., and Cornelius, J. M. (1986) Biochim. Biophys. Acta 849,16-24 50. Kojima, y., Hiyama, T., and Sakurai, H. (1987) in Progress in Photosynthesis Research (Biggins, J., ed) Vol. 11, pp. 57-60, Martinus Nijhoff, Dordrecht, The Netherlands 51. Malkin, R. (1984) Biochirn. Acta Biophys. 7 6 4 , 63-69 52. Morgan, T. V., Stephens, P. J., Devlin, F., Burgess, B.K., and Stout, C. D. (1985) FEBS Lett. 183, 206-210 53. Fish, L. E., Kuck, U., and Bogorad, L. (1985) in Molecular Biology of the Photosynthetic Apparatus (Steinback, K. E., Bonitz, S., Arntzen, C. J., and Bogorad, L., eds) pp. 111-120, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 54. A ~ R., ~~ ~ ~ , ~J., and ~ Vhnngird, ~ t T.~(1981) o ~ ~i ~ , ~ h i ~ . p h y ~Acta . 637,118-123 55. Palmer, G. (1975) in The Enzymes (Boyer, P. D., ed) Vol. 12, pp. 1-56, Academic Press, New York 56, Lundell, D. J., Glazer, A.N., Melis, A., and Malkin, R. (1985) J . Biol. Chem. 260,646-654 57. Takahashi, Y., Mitsui, A., Hase, T., and Matsubara, H. (1986) Proc. Natl.Acad.Sci. U. S. A. 83,2434-2437 58. McDermott, A. E., Yachandra, V. K., Guiles, R. D., Britt, R. D., Dexheimer, S. L., Sauer, K., and Klein, M. P. (1987) in Progress in Photosynthesis Research(Biggins, J., ed) Vol. I, pp. 249-252, Martinus Nijhoff, Dordrecht, The Netherlands 59. Golbeck, J. H., McDermott, A. E., Jones, w. K., and Kufiz, D. M. (1987) Biochirn. Biophys. Acta 891,94-98 60. Bonneiea, J. R.7 and Evans, M. c. w. (1984) Biochim. BioPhYs. Acta 767,153-159 61.d Kirsch, , W., Seyer, P., and Herrmann, R. G. (1986) Curr. Genet. 10,843-855 62. Lehmheck, J., Rasmussen, 0. F., Bookjans, G. B., Jepsen, B. R., Stummann, B. M., and Henningsen, K. W. (1986) Plant Mol. Biol. 7,3-10 63. Cushman, J. C., Hallick, R. B., and Price, C. A. (1987) in Progress in PhotosynthesisResearch (Biggins, J., ed) Vol. IV, pp, 667-670, Martinus Nijhoff, Dordrecht, The Netherlands 64. Bryant, D. A., Lorimier, R. D.,Guglielmi, G., Stirewalt, V. L., Cantrell, A,, and Stevens, S. E., Jr. (1987) in Progress in PhotosynthesisResearch (Biggins, J.,ed) Vol. IV, pp. 749-755, Martinus Nijhoff, Dordrecht, The Netherlands