* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Fatty acid
Size-exclusion chromatography wikipedia , lookup
Magnesium transporter wikipedia , lookup
Interactome wikipedia , lookup
Gene expression wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Signal transduction wikipedia , lookup
Peptide synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Western blot wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Metalloprotein wikipedia , lookup
Biosynthesis wikipedia , lookup
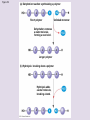
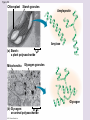
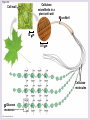
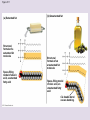
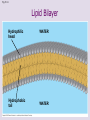
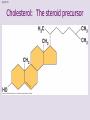
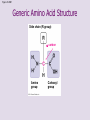
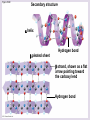
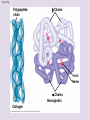
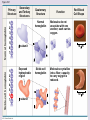
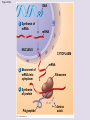
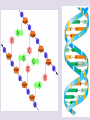
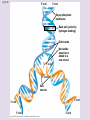
Chapter 5: The Structure and Function of Large Biological Molecules Four classes of organic molecules: • Carbohydrates • Lipids • Proteins • Nucleic Acids Organic molecules exhibit emergent properties Macromolecules are polymers • Polymers = large molecule consisting of smaller, nearly identical molecules joined together • Monomer = the small molecules used to build a polymer Figure 5.2 (a) Dehydration reaction: synthesizing a polymer 1 2 3 Short polymer Unlinked monomer Dehydration removes a water molecule, forming a new bond. 1 2 3 4 Longer polymer (b) Hydrolysis: breaking down a polymer 1 2 3 Hydrolysis adds a water molecule, breaking a bond. 1 2 3 4 Carbohydrates • Group includes sugars and polymers of sugars • Monomer = monosaccharide • Polymer = polysaccharide Monosaccharides • Simple sugars • Usually a multiple of a general molecular formula CH2O (e.g. C6H12O6) • Contain hydroxyl and carbonyl functional groups • Carbon skeletons vary in length from 3 to 7 carbons • Important as fuel for cells Figure 5.3 Aldoses (Aldehyde Sugars) Ketoses (Ketone Sugars) Trioses: 3-carbon sugars (C3H6O3) Glyceraldehyde Dihydroxyacetone Pentoses: 5-carbon sugars (C5H10O5) Ribose Ribulose Hexoses: 6-carbon sugars (C6H12O6) Glucose Galactose Fructose Figure 5.4 1 2 6 6 5 5 3 4 4 5 1 3 6 (a) Linear and ring forms 6 5 4 1 3 2 (b) Abbreviated ring structure 2 4 1 3 2 Disaccharides • Two monosaccharides joined together • Dehydration reaction • Examples: • Maltose (glucose + glucose) • Sucrose (glucose + fructose) Figure 5.5 1–4 glycosidic 1 linkage 4 Glucose Glucose Maltose (a) Dehydration reaction in the synthesis of maltose 1–2 glycosidic 1 linkage 2 Glucose Fructose (b) Dehydration reaction in the synthesis of sucrose Sucrose Polysaccharides • Long chains of monosaccharides • Function in energy storage and/or structure • Starch: energy storage in plants • Glycogen: energy storage in animals • Cellulose: structural in plants Figure 5.6 Chloroplast Starch granules Amylopectin Amylose (a) Starch: 1 m a plant polysaccharide Mitochondria Glycogen granules Glycogen (b) Glycogen: 0.5 m an animal polysaccharide Figure 5.7 (a) and glucose ring structures 4 1 4 Glucose Glucose 1 4 (b) Starch: 1–4 linkage of glucose monomers 1 1 4 (c) Cellulose: 1–4 linkage of glucose monomers Figure 5.8 Cellulose microfibrils in a plant cell wall Cell wall Microfibril 10 m 0.5 m Cellulose molecules Glucose monomer Lipids • Not a true polymer • Three categories 1. Fats/Oils (triglycerides) 2. Phospholipids 3. Waxes Figure 5.10 Fatty acid (in this case, palmitic acid) Glycerol (a) One of three dehydration reactions in the synthesis of a fat Ester linkage (b) Fat molecule (triacylglycerol) Figure 5.11 (a) Saturated fat Structural formula of a saturated fat molecule Space-filling model of stearic acid, a saturated fatty acid (b) Unsaturated fat Structural formula of an unsaturated fat molecule Space-filling model of oleic acid, an unsaturated fatty acid Cis double bond causes bending. Hydrophobic tails Hydrophilic head Figure 5.12 Choline Phosphate Glycerol Fatty acids Hydrophilic head Hydrophobic tails (a) Structural formula (b) Space-filling model (c) Phospholipid symbol Fig. 5-14 Lipid Bilayer Hydrophilic head Hydrophobic tail WATER WATER Waxes • One fatty acid connected to a glycerol • More hydrophobic than fats Fig. 5-15 Cholesterol: The steroid precursor Proteins • Function determined by 3-D shape (important!) • Monomer = amino acid • Polymer = polypeptide (protein = functional) Figure 5.15_a Proteins: Categorized based on function Enzymatic proteins Defensive proteins Function: Selective acceleration of chemical reactions Example: Digestive enzymes catalyze the hydrolysis of bonds in food molecules. Function: Protection against disease Example: Antibodies inactivate and help destroy viruses and bacteria. Antibodies Enzyme Virus Bacterium Storage proteins Transport proteins Function: Storage of amino acids Function: Transport of substances Examples: Hemoglobin, the iron-containing protein of vertebrate blood, transports oxygen from the lungs to other parts of the body. Other proteins transport molecules across cell membranes. Examples: Casein, the protein of milk, is the major source of amino acids for baby mammals. Plants have storage proteins in their seeds. Ovalbumin is the protein of egg white, used as an amino acid source for the developing embryo. Transport protein Ovalbumin Amino acids for embryo Cell membrane Figure 5.15_b Hormonal proteins Receptor proteins Function: Coordination of an organism’s activities Example: Insulin, a hormone secreted by the pancreas, causes other tissues to take up glucose, thus regulating blood sugar concentration Function: Response of cell to chemical stimuli Example: Receptors built into the membrane of a nerve cell detect signaling molecules released by other nerve cells. High blood sugar Insulin secreted Normal blood sugar Receptor protein Signaling molecules Contractile and motor proteins Structural proteins Function: Movement Examples: Motor proteins are responsible for the undulations of cilia and flagella. Actin and myosin proteins are responsible for the contraction of muscles. Function: Support Examples: Keratin is the protein of hair, horns, feathers, and other skin appendages. Insects and spiders use silk fibers to make their cocoons and webs, respectively. Collagen and elastin proteins provide a fibrous framework in animal connective tissues. Actin Myosin Collagen Muscle tissue 100 m Connective tissue 60 m Figure 5.UN01 Generic Amino Acid Structure Side chain (R group) carbon Amino group Carboxyl group Figure 5.16 Nonpolar side chains; hydrophobic Side chain (R group) Glycine (Gly or G) R group (side chain) properties Alanine (Ala or A) Methionine (Met or M) Isoleucine (Ile or I) Leucine (Leu or L) Valine (Val or V) Phenylalanine (Phe or F) Tryptophan (Trp or W) Proline (Pro or P) Polar side chains; hydrophilic Serine (Ser or S) Threonine (Thr or T) Cysteine (Cys or C) Electrically charged side chains; hydrophilic Tyrosine (Tyr or Y) Asparagine (Asn or N) Glutamine (Gln or Q) Basic (positively charged) Acidic (negatively charged) Aspartic acid (Asp or D) Glutamic acid (Glu or E) Lysine (Lys or K) Arginine (Arg or R) Histidine (His or H) Figure 5.17 Peptide Bond Formation Peptide bond New peptide bond forming Side chains Backbone Amino end (N-terminus) Peptide bond Carboxyl end (C-terminus) Four levels of protein structure • Primary • Secondary • Tertiary • Quaternary Figure 5.20a Primary structure Amino acids Amino end Primary structure of transthyretin Carboxyl end Figure 5.20c Secondary structure helix pleated sheet Hydrogen bond strand, shown as a flat arrow pointing toward the carboxyl end Hydrogen bond Figure 5.20b Tertiary structure Secondary structure Quaternary structure helix Hydrogen bond pleated sheet strand Hydrogen bond Transthyretin polypeptide Transthyretin protein Figure 5.20f Hydrogen bond Hydrophobic interactions and van der Waals interactions Disulfide bridge Ionic bond Polypeptide backbone Fig. 5-21g Polypeptide chain Chains Iron Heme Chains Hemoglobin Collagen Fig. 5-23 Denaturation Normal protein Renaturation Denatured protein Proteins can denature (lose shape) • High temperature • High/Low pH • Variation in salt concentration Figure 5.21 Sickle-cell hemoglobin Normal hemoglobin Primary Structure 1 2 3 4 5 6 7 Secondary and Tertiary Structures Quaternary Structure Function Molecules do not associate with one another; each carries oxygen. Normal hemoglobin subunit Red Blood Cell Shape 10 m 1 2 3 4 5 6 7 Exposed hydrophobic region Sickle-cell hemoglobin subunit Molecules crystallize into a fiber; capacity to carry oxygen is reduced. 10 m Nucleic Acids • Monomer = nucleotide • Polymer = polynucleotide (nucleic acid) • Blueprints for proteins • 2 types • DNA – Deoxyribonucleic Acid • RNA – Ribonucleic Acid Figure 5.25-3 DNA 1 Synthesis of mRNA mRNA NUCLEUS CYTOPLASM mRNA 2 Movement of mRNA into cytoplasm Ribosome 3 Synthesis of protein Polypeptide Amino acids Figure 5.26 5 end Sugar-phosphate backbone Nitrogenous bases Pyrimidines 5C 3C Nucleoside Nitrogenous base Cytosine (C) Thymine (T, in DNA) Uracil (U, in RNA) Purines 5C 1C 5C 3C Phosphate group 3C Sugar (pentose) Guanine (G) Adenine (A) (b) Nucleotide Sugars 3 end (a) Polynucleotide, or nucleic acid Deoxyribose (in DNA) (c) Nucleoside components Ribose (in RNA) Fig. 5-28 5' end 3' end Sugar-phosphate backbones Base pair (joined by hydrogen bonding) Old strands Nucleotide about to be added to a new strand 3' end 5' end New strands 5' end 3' end 5' end 3' end Figure 5.UN02