* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Protein Function and Classification
P-type ATPase wikipedia , lookup
Genetic code wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Molecular evolution wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
List of types of proteins wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Interactome wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein structure prediction wikipedia , lookup
Proteolysis wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein function and classification
Hsin-Yu Chang
www.ebi.ac.uk
Classifying proteins into families and identifying
protein homologues can help scientists to
characterise unknown proteins.
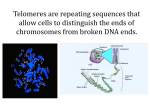
Greider and Blackburn discovered telomerase in 1984 and were awarded
Nobel prize in 2009. Which model organism they used for this study ?
2. Saccharomyces cerevisiae
3. Mouse
1. Tetrahymena
thermophila
4. Human
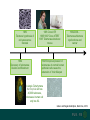
1989
Telomere hypothesis of
cell senescence
Szostak
1984
Discovery of telomerase
Greider and Blackburn
1995 Clone hTR
1995/1997 Clone hTERT
1997 Telomerase knockout
mouse
1999/2000…
Telomerase/telomere
dysfunctions and
cancer
1998 Ectopic expression of
telomerase in normal human
epithelial cells cause the
extension of their lifespan
A single Tetrahymena
thermophila cell has
40,000 telomeres,
whereas a human cell
only has 92.
Gilson and Ségal-Bendirdjian, Biochimie, 2010.
Can we identify human telomerase from
Tetrahymea protein sequence?
Let’s pretend that human telomerase has not been
identified and we only know the protein sequences
of Tetrahymena telomerase. How can we find the
human telomerase?
BLAST (Basic Local Alignment Tool)
: compares protein sequences to sequence databases
and calculates the statistical significance of matches.
BLAST
Advantages:
Drawbacks:
• Relatively fast
• User friendly
• sometimes struggle
with multi-domain
proteins
• Very good at
recognising similarity
between closely
related sequences
• less useful for weaklysimilar sequences
(e.g., divergent
homologues)
Using Tetrahymena telomerase protein sequences as a query in
BLAST, you will find a few human proteins that have very low identity.
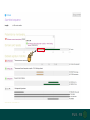
Tetrahymena and putative human telomerase (AAC51724.1) have
poor protein sequence match.
Can we presume this protein is a telomerase
homologue from humans? Can we find more
information about it before pursuing it further?
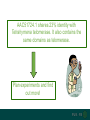
Search for protein signatures (such as
domains) in AAC51724.1
Telomerase
ribonucleoprotein
complex - RNA binding
domain
Reverse transcriptase
domain
AAC51724.1 shares 23% identity with
Tetrahymena telomerase. It also contains the
same domains as telomerase.
Plan experiments and find
out more!
But, where can we search for information
about the protein domains?
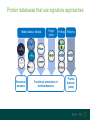
Protein databases that use signature approaches
Hidden Markov Models
Finger
prints
Profiles
Patterns
HAMAP
Structural
domains
Functional annotation of
families/domains
Protein
features
(sites)
Construction of protein signatures
• Construction of a multiple sequence alignment (MSA)
from characterised protein sequences.
• Modelling the pattern of conserved amino acids at
specific positions within a MSA.
• Use these models to infer relationships with the
characterised sequences
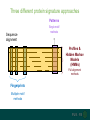
Three different protein signature approaches
Patterns
Sequence
alignment
Single motif
methods
Profiles &
Hidden Markov
Models
(HMMs)
Full alignment
methods
Fingerprints
Multiple motif
methods
Patterns
Patterns
Patterns are usually directed against functional sequence features such
as: active sites, binding sites, etc.
Sequence alignment
Motif
ALVKLISG
AIVHESAT
CHVRDLSC
CPVESTIS
Pattern sequences
[AC] – x -V- x(4) - {ED}
Regular expression
Pattern signature
PS00000
A conserved motif in tubulins
[SAG]-G-G-T-G-[SA]-G
Tubulin
signature
PDOC00199
Patterns
Advantages:
Drawbacks:
• Strict - a pattern with very
little variability and can
produce highly accurate
matches
• Simple but less
flexible
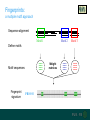
Fingerprints
Fingerprints:
a multiple motif approach
Sequence alignment
Motif 1
Motif 2
Motif 3
Define motifs
xxxxxx
xxxxxx
xxxxxx
xxxxxx
Motif sequences
Fingerprint
signature
PR00000
Weight
matrices
xxxxxx
xxxxxx
xxxxxx
xxxxxx
xxxxxx
xxxxxx
xxxxxx
xxxxxx
Motif 1
Motif 2
Motif 3
Telomerase signature (PR01365)
Motif 4
The significance of motif context
•
Identify small conserved regions in proteins
•
Several motifs characterise family
order
1
2
3
interval
Fingerprints
• Good at modeling the often small differences between closely
related proteins
• Distinguish individual subfamilies within protein families,
allowing functional characterisation of sequences at a high level
of specificity
Amino acids relatively well conserved across all chloride channel protein
family members
Amino acids uniquely conserved in chloride channel protein 3 subfamily
members.
Profiles & HMMs
Profiles & HMMs
Whole protein
Sequence alignment
Define coverage
Use entire alignment of
domain or protein family
Build model (Profile
or HMMs)
Profile or HMM
signature
Entire domain
xxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
Profiles
Start with a multiple
sequence alignment
Amino acids at each position
in the alignment are scored
according to the frequency
with which they occur
Scores are weighted
according to
evolutionary distance
using a BLOSUM matrix
• Good at identifying homologues
HMMs
Start with a multiple
sequence alignment
Amino acid frequency at each
position in the alignment and
their transition probabilities
are encoded
Insertions and deletions
are also modelled
Advantages
• Can model very divergent regions of alignment
• Very good at identifying evolutionarily distant homologues
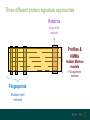
Three different protein signature approaches
Patterns
Single motif
methods
Profiles &
HMMs
hidden Markov
models
Full alignment
methods
Fingerprints
Multiple motif
methods
Fingerprints
Profiles &
HMMs
Patterns
hidden Markov
models
www.ebi.ac.uk/interpro
Hidden Markov Models
Finger
prints
Profiles
Patterns
HAMAP
Structural
domains
Functional annotation of
families/domains
Protein
features
(sites)
The aim of InterPro
Protein
sequences
Family entry: description,
proteins matched and more
information.
Domain entry: description,
proteins matched and more
information.
Site entry: description, proteins
matched and more information.
What is InterPro?
• InterPro is an integrated sequence analysis resource
• It combines predictive models (known as signatures)
from different databases
• It provides functional analysis of protein sequences by
classifying them into families and predicting domains and
important sites
Facts about InterPro
• First release in 1999
• 11 partner databases
• Add annotation to UniProtKB/TrEMBL
• Provides matches to over 80% of UniProtKB
• Source of >85 million Gene Ontology (GO) mappings to >24 million
distinct UniProtKB sequences
• 50,000 unique visitors to the web site per month> 2 million sequences
searched online per month. Plus offline searches with downloadable
version of software
InterPro signature integration process
• Signatures are provided by member databases
• They are scanned against the UniProt database to see which
sequences they match
• Curators manually inspect the matches before integrating the
signatures into InterPro
InterPro
curators
InterPro signature integration process
• Signatures representing the same entity are integrated together
• Relationships between entries are traced, where possible
• Curators add literature referenced abstracts, cross-refs to other
databases, and GO terms
http://www.ebi.ac.uk/interpro/
Search
using protein
sequences
Family
Type
InterPro entry types
Family
Proteins share a common evolutionary origin, as reflected in their
related functions, sequences or structure. Ex. Telomerase family.
Domain
Distinct functional, structural or sequence units that may exist in a
variety of biological contexts. Ex. DNA binding domain.
Repeats
Short sequences typically repeated within a protein. Ex. Tubulin
binding repeats in microtubule associated protein Tau.
Sites
PTM
Ex. Phosphorylation sites, ion binding sites, tubulin conserved site.
Active
Site
Binding
Site
Conserved
Site
Type
Name
Identifier
Contributing
signatures
Description
References
GO terms
Type
Identifier
Name
Relationships
Contributing
signatures
Description
References
InterPro family and domain relationships
Family relationships in InterPro:
Interleukin-15/Interleukin-21
family (IPR003443)
Interleukin-15
(IPR020439)
Interleukin-15
Avian
(IPR020451)
Interleukin-15
Fish
(IPR020410)
Interleukin-21
(IPR028151)
Interleukin-15
Mammal
(IPR020466)
Relationships
InterPro relationships: domains
Protein kinase-like
domain
Protein kinase
domain
Serine/threonine
kinase catalytic
domain
Tyrosine
kinase catalytic
domain
Gene Ontology
• Unify the representation of gene and gene product attributes across
species
• Allow cross-species and/or cross-database comparisons
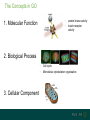
The Concepts in GO
•
•
1. Molecular Function
protein kinase activity
insulin receptor
activity
2. Biological Process
•
•
3. Cellular Component
Cell cycle
Microtubule cytoskeleton organisation
GO:0003677 DNA binding
GO:0003721 telomeric template RNA
reverse transcriptase activity
GO:0005634 Nucleus
Search
using
keywords
Summary
•
Protein classification could help scientists to gain information
about protein functions.
•
Blast is fast and easy to use but has its drawbacks.
•
Alternative approach: protein signature databases build
models (protein signatures) by using different methods
(patterns, fingerprints, profile and HMMs).
•
InterPro integrates these signatures from 11 member
databases. It serves as a sequence analysis resource that
classifies sequences into protein families and predicts
important domains and sites.
Why use InterPro?
• Large amounts of manually curated data
•
35,634 signatures integrated into 25,214 entries
•
Cites 38,877 PubMed publications
• Large coverage of protein sequence space
• Regularly updated
•
~ 8 week release schedule
•
New signatures added
•
Scanned against latest version of UniProtKB
Caution
• InterPro is a predictive protein signature database - results are
predictions, and should be treated as such
• InterPro entries are based on signatures supplied to us by our
member databases
....this means no signature, no entry!
And one more thing…..
We need your feedback!
missing/additional references
reporting problems
requests
EBI support page.
The InterPro Team:
Sarah
Hunter
Sebastien
Pesseat
www.ebi.ac.uk/interpro
Twitter: @InterProDB
Alex
Mitchell
Craig
McAnulla
Siew-Yit
Yong
Gift
Matthew
Maxim
Fraser Scheremetjew Nuka
Amaia
Sangrador
Hsin-Yu
Chang
Database
Basis
Institution
Built from
Focus
URL
Pfam
HMM
Sanger Institute
Sequence
alignment
Family & Domain
based on conserved
sequence
http://pfam.sanger.ac.uk/
Gene3D
HMM
UCL
Structure
alignment
Structural Domain
http://gene3d.biochem.ucl.a
c.uk/Gene3D/
Evolutionary
domain
relationships
http://supfam.cs.bris.ac.uk/
SUPERFAMILY/
Superfamily
HMM
Uni. of Bristol
Structure
alignment
SMART
HMM
EMBL Heidelberg
Sequence
alignment
Functional domain
annotation
http://smart.emblheidelberg.de/
Microbial Functional
Family Classification
http://www.jcvi.org/cms/rese
arch/projects/tigrfams/overv
iew/
TIGRFAM
HMM
J. Craig Venter Inst.
Sequence
alignment
Panther
HMM
Uni. S. California
Sequence
alignment
Family functional
classification
http://www.pantherdb.org/
PIRSF
HMM
PIR, Georgetown,
Washington D.C.
Sequence
alignment
Functional
classification
http://pir.georgetown.edu/pir
www/dbinfo/pirsf.shtml
PRINTS
Fingerprints
Uni. of Manchester
Sequence
alignment
Family functional
classification
http://www.bioinf.mancheste
r.ac.uk/dbbrowser/PRINTS/i
ndex.php
PROSITE
Patterns &
Profiles
SIB
Sequence
alignment
Functional
annotation
http://expasy.org/prosite/
HAMAP
Profiles
SIB
Sequence
alignment
Microbial protein
family classification
http://expasy.org/sprot/ham
ap/
ProDom
Sequence
clustering
PRABI : Rhône-Alpes
Sequence
alignment
Conserved domain
prediction
http://prodom.prabi.fr/prodo
m/current/html/home.php
Bioinformatics Center
Thank you!
www.ebi.ac.uk
Twitter: @emblebi
Facebook: EMBLEBI
YouTube: EMBLMedia
The BLOSUM (BLOcks SUbstitution Matrix) matrix is a
substitution matrix used for sequence alignment of
proteins. BLOSUM matrices are used to score alignments
between evolutionarily divergent protein sequences.
The BLOSUM (BLOcks SUbstitution Matrix) matrix is a
substitution matrix used for sequence alignment of
proteins. BLOSUM matrices are used to score alignments
between evolutionarily divergent protein sequences.