* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chapter 14
Eukaryotic DNA replication wikipedia , lookup
DNA repair protein XRCC4 wikipedia , lookup
Homologous recombination wikipedia , lookup
DNA profiling wikipedia , lookup
DNA replication wikipedia , lookup
DNA nanotechnology wikipedia , lookup
Microsatellite wikipedia , lookup
DNA polymerase wikipedia , lookup
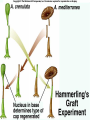
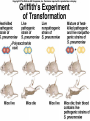
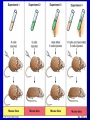
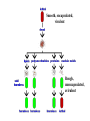
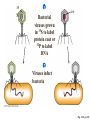
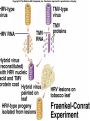
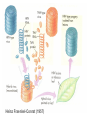
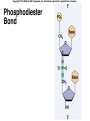
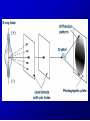
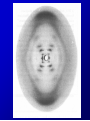
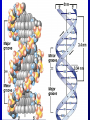
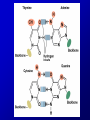
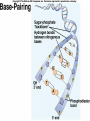
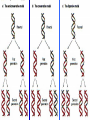
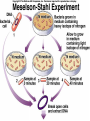
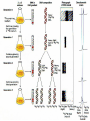
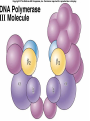
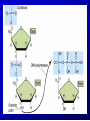
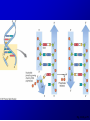
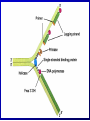
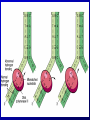
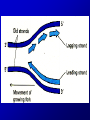
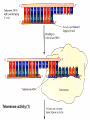
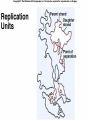
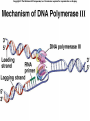
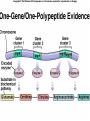
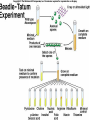
Lecture 26 - 27 DNA: THE GENETIC MATERIAL. Table 12-1, p. 263 Acetabularia mediterranea (Mermaid’s wineglass) Fig. A, p. 86 Fig. 12-1, p. 261 Smooth, encapsulated, virulent Rough, unencapsulated, avirulent 35 S 1 32 P Bacterial viruses grown in 35S to label protein coat or 32P to label DNA 2 Viruses infect bacteria Fig. 12-2, p. 262 3 Agitate cells in blender Agitate cells in blender 4 Separate by centrifugation Separate by centrifugation 35 S 5 32 P Bacteria in pellet contain 32P-labeled DNA 35S-labeled protein in supernatant Fig. 12-2, p. 262 Heinz Fraenkel-Conrat (1957) Friedrich Miescher (1844-1895)) Phoebius Levene (1869-1940) Erwin Chargaff (1905-2002) Rosalind Franklin The diagonal pattern of spots stretching from 11 to 5 and from 1 to 7 o’clock provides evidence for the helical structure of DNA. The elongated horizontal patterns at the top and bottom indicate that the purine and pyrimidine bases are stacked 0.34 nm apart and are perpendicular to the axis of the DNA molecule. X-ray diffraction image of DNA Rosalind Franklin (1952) X-ray crystallography Periodicity of 3.4 nm (= 10 nucleotides) Double helix Uniform width of 2 nm Bases are flat and perpendicular to the long axis of the molecule Lecture 28 - 29 DNA REPLICATION. Table 12-3, p. 270 DNA polymerase Fig. 12-10, p. 271 Telomerase activity (1) Telomerase activity (2) Fig. 12-15, p. 276 340 nm Fig. 12-13b, p. 274 The End.