* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PPT
Biology and consumer behaviour wikipedia , lookup
Genome (book) wikipedia , lookup
Gene desert wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genomic imprinting wikipedia , lookup
Designer baby wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Microevolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Population genetics wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Pathogenomics wikipedia , lookup
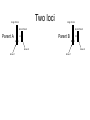
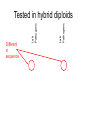
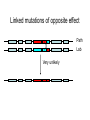
Variation in submergence tolerance QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. http://www.a2mediagroup.com/data/images/news/categories/riceplant.jpg Linkage mapping (quantitative) me™ and a ed) decompressor see this picture. intolerant tolerant Fine-mapping Finding the causative variant http://www.plantsci.cam.ac.uk/Haseloff/SITEGRAPHICS/Agrotrans.GIF Transgenic test QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Transgenic test QuickTime™ and a Uncompressed) decompressor needed to see this picture. http://homepages.strath.ac.uk/~dfs99109/BB310/CFTRgene.jpg Coding variants Fig. 2B Single-locus: AA x BB AB (F1) AB x AB AA AB BA BB (F2) 1 locus, incomplete dominance AB BA AA BB “Effect of having a B” 25% 50% 25% AA AB BA BB Two loci long chrom short chrom locus 2 locus 1 Two loci long chrom short chrom Parent A A A A A locus 2 locus 1 Two loci long chrom long chrom short chrom Parent A short chrom Parent B A A A A B B locus 2 locus 1 B B locus 2 locus 1 Two loci long chrom long chrom short chrom Parent A short chrom AA/AA x BB/BB A A Parent B A A B B locus 2 locus 1 B B locus 2 locus 1 Two loci long chrom long chrom short chrom Parent A short chrom AA/AA x BB/BB A A Parent B A A B B locus 2 locus 1 B B locus 2 AB/AB (F1) locus 1 Two loci long chrom long chrom short chrom Parent A short chrom AA/AA x BB/BB A A Parent B A A B B locus 2 locus 1 B B locus 2 AB/AB Locus1/Locus 2 (F1) locus 1 Two loci long chrom long chrom short chrom Parent A short chrom AA/AA x BB/BB A A Parent B A A B B locus 2 locus 1 B B locus 2 AB/AB (F1) Locus1/Locus 2 AB/AB x AB/AB 16 possibilities (F2) locus 1 Two loci, incomplete dominance Two loci, incomplete dominance or BA Two loci, incomplete dominance or BA or BA Two loci, incomplete dominance How many squares of the Punnett square are represented here? or BA or BA Two loci, incomplete dominance Effect of B at locus 2 Two loci, incomplete dominance Effect of B at locus 2 Two loci, incomplete dominance 0 0.5 1.0 Phenotype QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. 1.5 Two loci, incomplete dominance * 0 What is the genotype? A. AA/AA B. AB/AA C. BB/AA D. AA/AB QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. 0.5 1.0 Phenotype 1.5 Two loci, incomplete dominance * 0 What is the genotype? A. AA/AB B. BB/AB C. AB/AB D. AA/BB QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. 0.5 1.0 Phenotype 1.5 Two loci, incomplete dominance * 0 How many F2’s have this genotype? A. 1/16 B. 2/16 C. 3/16 D. 4/16 QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. 0.5 1.0 Phenotype 1.5 Two loci, incomplete dominance Notation is different, trait is qualitative; math is the same. Fig. 3.17 2-locus interaction Leghorn QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. http://www.projects.roslin.ac.uk/chickmap/organism.html Junglefowl QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Two loci long chrom long chrom short chrom Parent J J J short chrom JJ/JJ x LL/LL J Parent L J L locus 2 locus 1 L L L locus 2 JL/JL (F1) Locus1/Locus 2 JL/JL x JL/JL 16 possibilities (F2) locus 1 2-locus interaction QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. 2-locus interaction QuickTime™ and a TIFF (Uncompressed) decompressor Effect are needed to see this picture. of J at locus 2 2-locus interaction QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Effect of J at locus 2 2-locus interaction QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Effect of J at locus 2 Effect of variation at locus 2 depends on genotype at locus 1: non-additive 2-locus interaction For example: locus 1 is polymorphism in a transcription factor gene that affects protein binding affinity, locus 2 is in a binding site QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Effect of J at locus 2 Effect of variation at locus 2 depends on genotype at locus 1: non-additive 2-locus interaction QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Effect of J at locus 2 Locus 2 is epistatic to locus 1: effects of locus 1 are masked in individuals with JJ or JL,LJ at locus 2 Locus 2 follows a dominance model: JJ and JL,LJ have the same phenotype, LL differs “The dominant allele of locus 2 does the masking” Other foibles in QTL mapping Pathogenic yeast QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Pathogenic yeast QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. How did YJM145 gain the ability to grow at body temperature? A model quantitative trait A model quantitative trait Lab parent A model quantitative trait Lab parent Pathogenic parent A model quantitative trait 100 progeny ≥ pathogenic parent Lab parent Pathogenic parent NO progeny as extreme as diploid hybrid QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. NO progeny as extreme as diploid hybrid QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. NO progeny as extreme as diploid hybrid Lab parent Pathogenic parent Diploid hybrid Did the mapping… Significant linkage to yeast chrom XIV and chrom XVI. Did the fine-mapping… QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Did the fine-mapping… QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Which gene is causative? Lab Pathogenic Lab Pathogenic Tested in hybrid diploids Lab Pathogenic Lab Pathogenic Tested in hybrid diploids Lab Pathogenic Lab Pathogenic Tested in hybrid diploids Lab Pathogenic Different in sequence Lab Pathogenic Tested in hybrid diploids One mutant gene… From pathogenic strain QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. One mutant gene… From pathogenic strain From lab strain QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. One mutant gene… From pathogenic strain So diploid with this gene from the pathogenic strain grows BETTER at From lab strain QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. high T. Two mutant genes… From pathogenic strain QuickTime™ and a lab strain From TIFF (Uncompressed) decompressor are needed to see this picture. Two mutant genes… From pathogenic strain QuickTime™ and a lab strain From TIFF (Uncompressed) decompressor are needed to see this picture. Again, diploid with this gene from the pathogenic strain grows BETTER at high T. Three mutant genes From lab strain From pathogenic strain QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Diploid with this gene from the pathogenic strain grows WORSE at high T! Three mutant genes From pathogenic strain From pathogenic strain From pathogenic strain QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Three mutant genes From pathogenic strain From pathogenic strain From pathogenic strain QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Three mutant genes From pathogenic strain From pathogenic strain From pathogenic strain Alleles from the same strain at different genes/loci can have different effects. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. No common coding alleles across strains QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. No common coding alleles across strains QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. And no expression differences of >3-fold… Three mutant genes in same “locus” QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Three mutant genes in same “locus” QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Three mutant genes So why did 0/900 haploid progeny have a phenotype as extreme as the diploid hybrid? NO progeny as extreme as diploid hybrid Lab parent Pathogenic parent Diploid hybrid NO progeny as extreme as diploid hybrid QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Lab parent Pathogenic parent QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Diploid hybrid Linked mutations of opposite effect Path Linked mutations of opposite effect Path Lab Linked mutations of opposite effect Path Lab Very unlikely “Multiple linked loci”