* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Inferring genetic regulatory logic from expression data
Biology and consumer behaviour wikipedia , lookup
Genomic imprinting wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome (book) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Computational phylogenetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
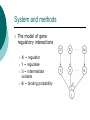
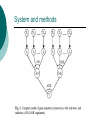
Inferring genetic regulatory logic from expression data Prepared by Chunhui CAI Logic gates Working principles of cis-regulatory elements ‘OR’ logic ‘AND’ logic ‘NOR’ logic ‘OR-NOR’ logic Bayesian network A probabilistic model Use probability as a means to express uncertainty about modeling variables and their dependencies. System and methods The model of gene regulatory interactions Xi ~ regulator Y ~ regulatee Ii ~ intermediate variable θi ~ binding probability System and methods System and methods System and methods ‘OR’ model System and methods ‘OR-NOR’ model System and methods Rules Gene expression values ~ two states 0 – not expressed 1 – expressed Type of regulation Simultaneous Time delay Result Regulatory interactions of 20 genes of S.cerevisiae. The full arcs represent activatory regulation, the dashed arcs represent inhibitory regulation. The relationship between genes regulating one common gene is described by ‘OR’function. Discussion Probabilistic or deterministic Either ~ Fitting of the expression data into state value (simply two states(0,1)?) Or ~ Determining the activation/repression logic (Later) How good is our clustering method (need to improve and more expression data should be involved)