* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Régulation de SRY - Département de biologie
List of types of proteins wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene desert wikipedia , lookup
Community fingerprinting wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genome evolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
RNA silencing wikipedia , lookup
Non-coding RNA wikipedia , lookup
Molecular evolution wikipedia , lookup
Gene expression wikipedia , lookup
Gene regulatory network wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Promoter (genetics) wikipedia , lookup
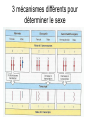
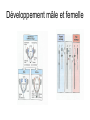
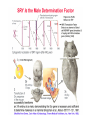
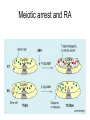
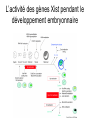
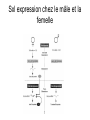
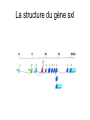
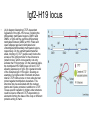
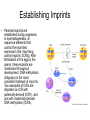
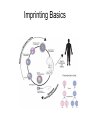
3 mécanismes différents pour déterminer le sexe Développement mâle et femelle Expression de la protéine SRY La séquence de SRY Régulation de SRY Meiotic arrest and RA Spermatogenèse murine TRD Chromosome 17 TRD Distorters et Responder des tmutants TRD Smok Kinase et TRD Tortoiseshell cat/ Ecaille de tortue Clonal selection • Schematic representation of XCI in female somatic cells. In normal conditions, the ratio of the two cell types (carrying the active and the inactive X chromosomes) is approximately 50:50, but in females with Xlinked dominant disorders this ratio can be different because of a disadvantage for cells expressing a mutant X-linked allele. Divergence from the 50:50 ratio, known as skewing of XCI, can be different in various tissues and in different developmental stages, and can vary among individuals, causing a variable severity of the phenotype observed. For disorders such as MLS, OFCD, ODPD and IP, affected females usually have totally skewed patterns of XCI, in favour of an active wild type X chromosome. Cell selection usually affects only those cell lines in which the disease gene is expressed [43].. Activité des chromosomes X pendant le développement Conservation du gène Xist X inactivation centre Xist inactivation complèxe Etapes moléculaires pendant l’inactivation du chromosome X Modifications des histones du gène Xist L’activité des gènes Xist pendant le développement embryonnaire Histone Code Counting chromosomes Mâle ou femelle chez la drosophile? Sxl et les 2 promteurs p et m Sxl expression chez le mâle et la femelle La structure du gène sxl L’épissage de sxl Contrôle de l’épissage par tra et msl-2 Transfert des pronoyau Disomy et développement Imprinting Models Igf2-H19 locus • (A) A diagram illustrating CTCF-dependent regulation of the Igf2–H19 locus, including the differentially methylated regions, DMR1 and DMR2, of Igf2 and the germline differentially methylated domain (DMD) at H19. Filled and open lollipops represent methylated and unmethylated differentially methylated regions, respectively. On the unmethylated maternal allele, binding of CTCF (purple oval) blocks the access of the Igf2 promoter to the enhancers (small circles), which consequently can only activate the H19 promoter. On the paternal allele, the methylated H19 DMD does not bind CTCF, allowing expression of Igf2. (B) A general model of the maternal Igf2–H19 region, showing an example of a higher-order chromatin structure where CTCF binds at one or more sites but can protect against methylation elsewhere. This structure may be associated with the nuclear matrix and involve proteins in addition to CTCF. Tissue-specific variation in higher-order structure could be due to different CTCF-dependent cis elements forming the base of the loop or different proteins acting in trans. Imprinted Genes • Genomic organization of two imprinted clusters. (a) The Igf2/H19 locus. The maternally inherited H19 gene expresses an untranslated RNA of unknown function when the nearby imprinting center (IC) is unmethylated, in which context, Igf2 is repressed. On the paternally derived allele, the IC is methylated (asterisks) and this represses H19, but enables Igf2 expression. (b) The UBE3A/UBE3A-ATS locus. The maternally derived alleles of the UBE3A and ATP10C genes are expressed, but the UBE3A antisense (UBE3A-ATS), SNURF/SNRPN, MKRN3, NDN and MAGEL2 genes are silent and the IC just upstream of the SNRPN promoter is methylated (asterisks). When paternally inherited, the upstream MKRN3, NDN and MAGEL2 genes are expressed, as is the long noncoding antisense RNA, UBE3A-ATS. The UBE3A and ATP10C genes are repressed. The c15orf2 gene, which lies inside this imprinted cluster, is not subject to imprinting. Igf2r/Air • Imprinting mechanisms across the Igf2r/Air locus. (a) The maternal allele. The Igf2r, Slc22a1 and Slc22a3 genes are all expressed only from the maternally inherited allele (orange boxes). Air is silent and the imprint control region (Region 2, red box) adjacent to the Air promoter is methylated (asterisk). Genes not subject to imprinting are shown as blue boxes. (b,c) The paternal allele. (b) In a one-step model, Air RNA (red line) associates with repressor proteins (purple ovals) to form silencing complexes that associate with sequences within or around genes subject to imprinting (orange boxes). In the case of Igf2r, promoter methylation (asterisks) could be a consequence of its silent status. (c) In a twostep model, Air transcription and/or Air RNA induces Igf2r promoter methylation (asterisks). The Igf2r silencing induced by this promoter methylation then spreads (dashed grey line and arrows) to other genes (orange boxes) in some tissues and at certain developmental stages Prader-Willi • Molecular classes of Prader–Willi and Angelman syndrome. Each genetic event is parental-specific in PWS and AS. Deletions and imprinting mutations occur with equal frequency in both syndromes, whereas UPD is more common in PWS than AS because of higher rates of maternal nondysjunction. The gene mutation class in AS appears to be lacking in PWS, which probably indicates that PWS represents a contiguous gene syndrome. Abbreviations: hatched chromosome, non-chromosome 15 in rare balanced translocations; M, maternal (red); M(P), maternal inheritance of imprinting center (IC) mutations with a fixed paternal epigenotype (horizontal lines); P, paternal (blue); P(M), paternal inheritance of IC mutations with a fixed maternal epigenotype; UPD, uniparental disomy. Angelman Prader Willi Establishing Imprints • Parental imprints are established during oogenesis, or spermatogenesis, at sequence elements that control the imprinted expression (the ‘imprinting control regions’ [ICRs]). After fertilisation of the egg by the sperm, these imprints are maintained throughout development. DNA methylation (lollypops) is the most consistent hallmark of imprints. Two examples of ICRs are depicted: an ICR with paternally-derived (ICR1), and one with maternally-derived DNA methylation (ICR2). Igf2-H19 • The establishment and maintenance of DNA methylation at the ICR of the Igf2–H19 locus. This ICR acquires methylation (lollypops) during late spermatogenesis. In the female germ line, by contrast, the ICR is protected against methylation by the zincfinger protein CTCF. After fertilisation, and throughout development, the maternal allele (M, pink) continues to be protected against methylation by CTCF binding. CTCF binding onto the maternal chromosome creates a chromatin boundary that prevents interaction between the Igf2 gene and enhancers (ovals) located downstream of H19. The methylation on the paternal allele (P, blue) is maintained throughout development and prevents CTCF binding. On the paternal chromosome, the Igf2 promoters can therefore interact with the enhancers, but here the H19 gene is repressed because of the ICR methylation spreading into the nearby H19 promoter. DNA methylation/ICR • Sequence elements are important for the sex-specific establishment of DNA methylation at ICRs. (a) At the ICR controlling the mouse Rasgrf1 locus, the male-specific establishment of methylation requires the presence of neighbouring repeat sequences (open arrows). (b) The ICR at the SNRPN gene regulates imprinting along a large chromosomal domain, which is deregulated in the Prader–Willi syndrome (PWS). For the acquisition of methylation at the ICR in the female germ line, a region located >30 kb further upstream (the AS region) is essential. ICRs • At the ICRs controlling (a) the Igf2r and (b) the Kcnq1 domains, non-coding RNAs are produced from the unmethylated paternal allele. These non-coding RNAs (Air at Igf2r, and Lit1 at Kcnq1) could be involved in the paternal repression along these domains, which extends furthest in the placenta at both the domains. This developmentally-regulated silencing mechanism bears similarities with (c) imprinted X-chromosome inactivation in the mouse placenta, which is brought about by ‘coating’ of the paternal X-chromosome by the noncoding Xist transcript, and subsequent recruitment of chromatin-modifying complexes. Imprinting Basics