* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Twenty-five years of the nucleosome Kornberg and Lorch 1998, Cell
Minimal genome wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Transcription factor wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
X-inactivation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Point mutation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Primary transcript wikipedia , lookup
Neocentromere wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
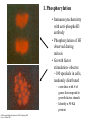
Twenty-five years of the nucleosome Kornberg and Lorch 1998, Cell 98: 285. HATs HDACs Important point• P289- Although acetylation of histone tails may counteract condensation of nucleosomes in chromatin fibers, it is unlikely to disrupt the structure of the core particle for transcription • Why? Tails are outside of the core, make little contribution to overall structure • Chromatin remodeling enzymes are likely responsible for nucleosome disruption Chromatin remodeling • SWI/SNF proteins are chromatin remodelers • These disrupt nucleosome in an ATPdependent fashion (ATPase activity) • Models– displacement – octamer sliding Histones- modifications and function Michael Hagmann 1999 Science 285:1203 • A. Phosphorylation of histones • two opposing functions reported » opening chromatin » condensing chromatin (cell division) I. Phosphorylation • Immunocytochemistry with anti-phosphoH3 antibody • Phosphorylation of H3 observed during mitosis • Growth factor stimulation- observe ~100 speckels in cells, randomly distributed – correlates with # of genes that respond to growth factor stimuli. – Identify a 90 Kd protein Cellular and Molecular Genetics BLA510 Spring 2001 Gary A. Bulla, PhD B. Coffin Lowry syndrome- mental retardation and a defect in growth factor response • mutation identified in in Rsk-2 gene •Immunocytochemistry with anti-phosphoH3 Ab -no speckles observed Growth factor Receptor MAP kinase signal transduction pathway Thus, Rsk-2 mutation prevented H3 phosphorylation Ras Raf (MAPKKK) MEK (MAPKK) ERK RSK-2 H3 P H3 ( MAPK) ExperimentInduce cells with growth factors, Crosslink DNA+ proteins, then immunoprecipitate with anti-Phospho-H3 Ab Most genes H3 Crosslink, nuclease H3 c-fos P H3 P DNA H3 Immunoprecipitate with anti-Phospho-H3 Thus, known growthresponse genes are bound by histones with phosphorylated H3 Agarose gel Southern Probe with labeled c-fos DNA C. Role in phosphorylation in cell division 1. Tetrahymena # copies of chromosomes macronucleus 90 micronucleus 2 Mode of replication in cell division Pinching off Normal mitosis Affect of H3 mutation at phosphorylation site none abnormal condensation chromosome loss Tetrahymena (a protozoan) Genetics, Russell, p6. Tetrahymena- Histone H3 phosphorylation occurs only in mitotic micronuclei Macronuclei Micronuclei Mitotic (football shape) 2. Immunocytochemistry- observe phospho-H3 throughout chromosomes during cell division Thus, this must play a role is chromosome condensation during mitosis 3. Models1. Phosphorylation + acetylation allows activation of gene expression, depending on context 2. Phospho-H3 loosens chromatin, enhancing transcription factor binding or mitotic factor binding II. Methylation CARM-1 - • activates transcription (coactivator) • methylates proteins • inactivation of methylation activity - lose transcriptional activation • methylates histone H3 in vitro • what are CARM-1 targets?? coactivator Steroid hormone receptor CARM-1 p160 Me H3 +1 TATAA III. Acetylation- Bromodomain -100 AA found in ~30 chromatin associated proteins (inc. HATs) - may be binding motif for actetylated histones Acetylated lysine IV. Other modifications- ubiquitination, glycosylation Methylation, phosphorylation and acetylation of histones OFF Histone H3 ON Bromodomain of a HAT Chromodomain of a chromatin remodeler Science 292:65, 2001 Is there a “Histone Code”? • Definition- “Covalent modifications of histones constitute an intricate pattern that creates a docking surface with which the modules of other proteins can interact” Shelley Berger, Wistar Institute Science 292:65, 2001