* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Linkage analysis
Microevolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genomic library wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Public health genomics wikipedia , lookup
Pharmacogenomics wikipedia , lookup
SNP genotyping wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Population genetics wikipedia , lookup
Genetic drift wikipedia , lookup
Genome-wide association study wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Dominance (genetics) wikipedia , lookup
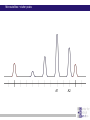
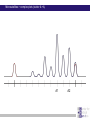
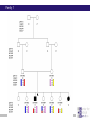
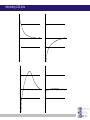
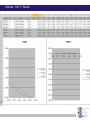
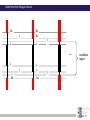
Chapter 6 Linkage analysis Jan Hellemans Aims Interprete microsatellite results Add genotypes to pedigrees Create pedigree and genotype files Calculate and interprete LOD-scores Delineate linkage intervals Basic principles of linkage analysis Analyze other types of markers Association studies Learn how to work with specific pedigree programs Starting linkage analysis Preparations Clearly define the phenotype If not specific enough than you may analyze different disorders that can map to different genomic loci Find suitable families larger is better more patients is better Collect genomic DNA from as much family members as possible Determine the type of inheritance Calculate the power to proove linkage with the available material (SLink – not part of this course) Linkage analysis types Directed linkage analysis Evaluate linkage at a specific locus such as a candidate gene Common approach: evaluate an intragenic, 5’ and 3’ marker Genome wide linkage analysis Screen for linkage for markers spread across the entire genome Microsatellites: ~400 markers spaced at about 10cM SNP’s: 500k SNP array Homozygosity mapping Screen only affected individuals in inbred families Select homozygous markers Very efficient technology Exercise – Part 1 2 inbred families with a recessive disorder With a homozygosity mapping based on 500k SNP arrays 2 candidate regions could be identified Chromosome 4 Patient 1 homozygous for 6.052Mb - 14.488Mb 21.008Mb – 37.477Mb Patient 2 homozygous for 11.186Mb – 37.219Mb 40,000 35,000 30,000 25,000 20,000 15,000 Task: find microsatellite markers to confirm linkage 10,000 5,000 1 2 Find additional flanking markers Find physical position of marker in NCBI > UniSTS NCBI map viewer: http://www.ncbi.nlm.nih.gov/mapview/ Go to Homo sapiens and to the right chromosome Maps & options: show DeCode, Généthon & Marshfield (genetic maps) Genes Set region: e.g. 2Mb up- and downstream of your marker Click ‘Data as table view’ Click on STS behind a marker to see its details Select markers that locate to only 1 genomic location have a PCR product with an extended size range one size not polymorphic Exercise – Part 1 > possible solution Markers in 1st candidate region D4S3017 (21.078Mb) D4S3044 (25.189Mb) D4S1618 (33.857Mb) D4S3350 (33.857Mb) D4S2988 (36.889Mb) Markers in 2nd candidate region D4S1582 (10.311Mb) D4S2906 (12.321Mb) D4S2944 (13.141Mb) D4S1602 (14.059Mb) D4S2960 (15.437Mb) Order primers & analyze them on all family members Analyzing microsatellite data Microsatellites > basics Repeats of short sequences (e.g. 2bp) NNNNAC(AC)nACNNNN Number of repeats is variable (instable sequence) Number of repeats determines the allele Number of repeats corresponds to specific length of PCR product: allel 1: NNNNACACACACACNNNN allel 2: NNNNACACACACACACNNNN allel 3: NNNNACACACACACACACNNNN ... (5*AC 18bp) (6*AC 20bp) (7*AC 22bp) Determine length to know the allele (sequencer) Microsatellites > basics Microsatellites > determine size Use internal size standard (other color) 220bp 230bp 225bp Microsatellites > heterozygotes 220bp 230bp 223bp 225bp Microsatellites > stutter peaks Repeats are difficult to copy polymerase slips Some amplicons have 1 repeat less a few even loose multiple repeats Small repeats are more prone to slippage and show more pronounced stutter peaks Largest product is the correct one Distance between peaks = length of a repeat Microsatellites > stutter peaks allelic peak 1st stutter peak 2nd stutter peak Microsatellites > stutter peaks Allelic peaks are the heighest Stutter peaks are lower A1 A2 Microsatellites > stutter peaks A1 A2 Microsatellites > +A peaks Taq polymerase tends to add an extra A at the 3’ end Variable degree of products with or without this extra A Do not confuse with stutter peaks (only 1bp difference) allelic peak allelic peak + A 1st stutter peak 1st stutter peak + A 2nd stutter peak 2nd stutter peak + A Microsatellites > complex plots (stutter & +A) A1 A2 Microsatellites > mutliplex Combine multiple markers in a single analysis ($$$) Different size range Multicolor Commercial kits: e.g. 16 markers / lane Microsatellite plots examples Genotyping pedigrees Genotyping pedigrees Screen one or multiple markers for some or all family members For every marker: Make a list of all occuring allele sizes Due to technical variation on sizing the same allele can have a slightly different size in different measurements (-0.4bp _ +0.4bp). Give all alleles within this range the same allele number Add the allele numbers to the pedigree at the corresponding individual/marker combination Find the wright phase Advanced software like GeneMapper can generate tables with allele numbers for every sample / marker Advanced pedigree programs like Progeny can store genotype information for family members Verify inheritance Exercise – Part 2 Genotype 3 markers in all available individuals of 2 families Pedigrees & microsatellite plots in ExercisePart2-GenotypingData.pdf Add allele numbers for the 3 markers to the pedigree Interprete the genotyped pedigrees: linked? Family 1 Family 2 Exercise – Part 2 > Conclusions D4S1582 Mendelian error can not be interpreted D4S2944 Linked D4S3017 Not-linked: unaffected individuals with the same genotype as a patient Calculate LOD scores EasyLinkage EasyLinkage = UI for linkage analysis http://genetik.charite.de/hoffmann/easyLINKAGE/index.html#start Bioinformatics. 2005 Feb 1;21(3):405-7 PMID: 15347576 Bioinformatics. 2005 Sep 1;21(17):3565-7 PMID: 16014370 Interface for many linkage analysis programs Input Pedigree file (linkage format) Genotype file(s) Marker information (already provided for popular markers) Settings Pedigree file Naming requirements for EasyLinkage: p_xxx.pro e.g. p_SMMD.pro Format: Tab delimited text file 1 individual per row Columns: 1 family ID 2 person ID 3 father ID 4 mother ID 5 sex (1=male, 2=female, 0=unknown) 6 affection status (1=unaffected, 2=affected, 0=unknown) 7 DNA availability (optional, relevant for power calculations) 8 liability class (to be provided if multiple liability classes are used) Genotype files Person ID’s have to match exactly with those provided in the pedigree file Naming requirements for EasyLinkage: MarkerName_xxx.abi e.g. D1S1609_SMMD.abi Format: Tab delimited text file 1 individual per row Columns (for microsatellite based analysis): 1 marker (same as in file name and matching a marker in an available marker set) 2 custom information (content doesn’t matter, but column must be present) 3 individual ID (match person ID in pedigree file) 4 & 5 genotypes for 2 alleles (unknown=0) Marker information Contains information on the chromosome and position of every marker Already available for a number of commercial SNParrays and for the microsatellite markers from Genethon Marshfield DeCode Custom marker sets can be created (see manual) EasyLinkage settings Choose a program: FastLink Parametric, single-point SuperLink Parametric, single-/multipoint SPLink Nonparametric, single-point Genehunter Nonpara-/parametric, single-/multipoint Genehunter Plus Nonpara-/parametric, single-/multipoint Genehunter MOD Nonpara-/parametric, single-/multipoint Genehunter Imprinting Nonpara-/parametric, single-/multipoint GeneHunter TwoLocus Parametric, two-locus, single-/multipoint Merlin Nonpara-/parametric, single-/multipoint SimWalk Nonparametric, single-/multipoint Allegro Nonpara-/parametric, single-/multipoint & simulation, single/multi-point PedCheck Mendelian error check FastSLink Simulation, single-/multi-point EasyLinkage settings Parametric <-> non-parametric Single point <-> multipoint Frequency of the disease allele Penetrance vectors (wt/wt, wt/mt, mt/mt) Standard dominant: 0 1 1 Standard recessive: 0 0 1 Reduced penetrance: replace 1 by penetrance (e.g. 0.9) Phenocopy: replace 0 by percentage of phenocopy (e.g. 0.1) Example: 0.01 0.9 0.99 1% chance to show a similar phenotype despite a normal genotype 90% chance to show the phenotype when 1 mutant allele (dominant with incomplete penetrance) 99% likelihood to present with the phenotype if both alleles are mutant Evaluate calculated LOD-scores Maximum LOD-scores can be seen in EasyLinkage Details about LOD-scores at different recombination fractions can be fount in text files generated by EasyLinkage process in Excel (generate graphs, ...) Standard rules for LOD-scores >3 significant linkage 2<LOD<3 suggestive linkage -2<LOD<2 uninformative <-2 significant absence of linkage Interpreting LOD plots 5 5 4 4 3 3 2 2 1 1 0 0 0 0,1 0,2 0,3 0,4 0,5 -1 -1 -2 -2 -3 -3 -4 -4 -5 -5 5 5 4 4 3 3 2 2 1 1 0 0 0,1 0,2 0 0,1 0,2 0,3 0,4 0,5 0 0 0,1 0,2 0,3 0,4 0,5 -1 -1 -2 -2 -3 -3 -4 -4 -5 -5 0,3 0,4 0,5 Exercise – Part 3 Generate one pedigree file containing all family members of both families (use Global ID’s) Generate a genotype file for each of the tested markers Run SuperLink analysis with the right settings Evaluate results Exercise – Part 3 > Results Strengthen the evidence Analyze more family members Analyze more families Analyze flanking markers Look for more informative markers that result in higher LOD-scores A series of flanking markers allow for multipoint linkage analysis A series of linked markers gives more confidence (subjective) Flanking markers can also be used to fine-map the linkage interval Determine the linkage interval NL ? L NL NL L ? ... L ? NL L L ? NL candidate region Exercise 2: find the linkage interval Post linkage Create a list of all the genes within the linkage interval NCBI map viewer UCSC (also for non-coding RNA’s) Evaluate known gene functions for relevance to the investigated phenotype Sequence genes Start with those that seem the most relevant to the disorder Start with the coding regions Finding a mutation and proving its causality is the ultimate proof





















































![Department of Health Informatics Telephone: [973] 972](http://s1.studyres.com/store/data/004679878_1-03eb978d1f17f67290cf7a537be7e13d-150x150.png)