* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PPT - AePIC
G protein–coupled receptor wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Protein moonlighting wikipedia , lookup
Biochemistry wikipedia , lookup
Size-exclusion chromatography wikipedia , lookup
Protein design wikipedia , lookup
Protein domain wikipedia , lookup
Molecular evolution wikipedia , lookup
Homology modeling wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Western blot wikipedia , lookup
Circular dichroism wikipedia , lookup
List of types of proteins wikipedia , lookup
Interactome wikipedia , lookup
Protein structure prediction wikipedia , lookup
Implicit solvation wikipedia , lookup
Protein adsorption wikipedia , lookup
History of molecular evolution wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
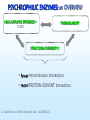
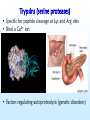
COMPARATIVE MOLECULAR DYNAMICS SIMULATIONS TO STUDY ENZYMATIC COLD ADAPTATION Luca De Gioia Molecular Modeling Laboratory, Department of Biotechnology and Biosciences, University of Milano-Bicocca, Italy PSYCHROPHILIC ORGANISMS • ARCTIC AND ANTARCTIC ½ Earth’s surface: oceans 1°C - 4°C Deep sea – 1°C to 4°C PSYCHROPHILIC ENZYMES: catalysis in extreme conditions rational design of biocatalysts and biotechnological applications Georlette et al, FEMS Microbiol. Rev., 28 (2004) 25. PSYCHROPHILIC ENZYMES: an OVERVIEW HIGH CATALYTIC EFFICIENCY at 0-30°C THERMOLABILITY STRUCTURAL FLEXIBILITY ? • fewer intramolecular interactions • more PROTEIN-SOLVENT interactions Georlette et al, FEMS Microbiol. Rev., 28 (2004) 25. Detailed INTRA-FAMILY structural comparisons COMPARATIVE and STATISTICAL INVESTIGATIONS • general features • overlooking subtle structural modifications A general theory of enzymatic cold adaptation cannot be formulated because… Cold adaptation in different families is most probably obtained by different EVOLUTIONARY STRATEGIES Gianese, G., Bossa, F., Pascarella, S., Proteins, 47 (2002) 236. Molecular dynamics Proteins are not rigid molecules • • • • Conformational changes Protein folding Molecular recognition (drug design) Ion transport The method is based on the Newton’s equation of motion: Fi mi ai Fi iV d 2 ri dV mi 2 dri dt •(Numerical) integration of the equation of motion yields a trajectory. •The average values of properties can be determined from the trajectory MD shortcomings • The integration step (dt) must be very small (1fs) [supercomputing] • The trajectory must be very long (to compute properties the simulation must pass through all possible states corresponding to the particular thermodynamic constraints) [supercomputing] MD protocol To properly sample the phase space: Multiple MD SIMULATIONS: Gromacs (50 ns, explicit solvent) MD protocol rmsd 1 N 2 ( r r ) i 0 N i1 Up to 10 ns •Rmsd (mainchain) N = number of atoms r = position; r0 = initial position •Protein gyration radius •Total and potential energy Elastases (serine protaeses) 3D STRUCTURE: 2 DOMAINS antiparallel β-type fold (12 β-strands and 3 α-helices). COLD-ADAPTED = atlantic salmon elastase (SE) MESOPHILIC = porcine elastase (PE) FUNCTIONAL SITES: catalytic triad (H57, D102 and S195) and specificity pocket Psychrophile/Mesophile comparison: primary sequences Primary sequence (PE and SE, ~ 210-250 aa ): 68.2% identity 76 amino acidic substitutions (45 completely unrelated aa). Maiale Bovina MerluzzoB Salmone Maiale Bovina MerluzzoB Salmone Maiale Bovina MerluzzoB Salmone VVGGTEAQRNSWPSQISLQYRSGSSWAHTCGGTLIRQNWVMTAAHCVDRELTFRVVVGEH VVGGTAVSKNSWPSQISLQYKSGSSWYHTCGGTLIKQKWVMTAAHCVDSQMTFRVVLGDH VVGGEDVRVHSWPWQASLQYKSGNSFYHTCGGTLIAPQWVMTAAHCIGSR-TYRVLLGKH VVGGRVAQPNSWPWQISLQYKSGSSYYHTCGGSLIRQGWVMTAAHCVDSARTWRVVLGEH **.* . ::** * *** .. : *.***:*: **:*****:. :** :* * NLNQ-NNGTEQYVGVQKIVVHPYWNTDDVAAGYDIALLRLAQSVTLNSYVQLGVLPRAGT NLSQ-NDGTEQYISVQKIVVHPSWNSNNVAAGYDIAVLRLAQSATLNSYVQLGVLPQSGT NMQDYNEAGSLAISPAKIIVHEKWD—-SSRIRNDIALIKLASPVDVSAIITPACVPDAEV NLNT-NEGKEQIMTVNSVFIHSGWNSDDVAGGYDIALLRLNTQASLNSAVQLAALPPSNQ .: : .:.:* *: *:*:::: . . : . :* ILANNSPCYITGWGLTRTNGQLAQTLQQAYLPTVDYAICSSSSYWGSTVKNSMVCAGGDG ILANNTPCYITGWGRTKTNGQLAQTLQQAYLPSVDYATCSSSSYWGSTVKTTMVCAGGDG LLANGAPCYVTGWGRLWTGGPIADALQQALLPVVDHAHCSRYDWWGSLVTTSMVCAGGDG ILPNNNPCYITGWGKTSTGGPLSDSLKQAWLPSVDHATCSSSGWWGSTVKTTMVCAGG-G :*.. **:**** *.* . *:*. : .: ** :*** : . *:*.** * Comparative molecular dynamics (MD) simulations Molecular flexibility is difficult to estimate experimentally but possibly crucial to understand cold-adaptation Comparative MD simulations of proteins •TIME-EVOLUTION of MOLECULAR PROPERTIES • evaluation of PROTEIN FLEXIBILITY Analysis of MD trajectories Secondary Structure (SS) content Hydrogen bonds Solvent accessible surface Psychrophile/Mesophile comparison: flexibility • Identification of regions characterized by different flexibility in SE and PE Root mean square fluctuation (Rmsf) profiles: highlight STRUCTURAL FLEXIBILITY. rmsf 1 N 2 ( r r ) i N i1 N = number of atoms r = position; <r> = average position Psychrophile/Mesophile comparison Different RMSF Amino acid COMPOSITION LOCALIZATION on the 3D structure Differences that could be related to cold adaptation SE PE Insight obtained by MD simulations • COLD-ADAPTED ELASTASES: localized flexibility (proximity of catalytic site/specificity pocket). • MESOPHILIC ELASTASES: scattered flexibility (far from protein functional sites). Design of “wet” experiments: site-directed mutagenesis Papaleo, E., Fantucci, P., De Gioia L., J. Chem. Theory Comput., 1 (2005) 1286. Trypsins (serine proteases) • Specific for peptide cleavage at Lys and Arg sites • Bind a Ca2+ ion • Factors regulating autoproteolysis (genetic disorders) MD investigation Role of Ca2+ in structure stabilization and autolysis? - The region K60-R117 (including the Ca2+ binding loop) can be a target for autolysis. - Ca2+ has been proposed to induce an autolysis-resistant conformation - Autolysis in fish trypsins is less Ca2+ dependent • Bovine and salmon trypsins • Apo and holo forms • Multiple MD simulations: ~ 200 ns Investigation of autoproteolysis sites • Effects due to Ca removal Flexibility of R117 and K188 is enhanced in BT Insight from MD simulations • Ca2+ removal increases the flexibility of residues forming the binding site, but… • …it also leads to enhanced flexibility in remote regions • Ca2+ affects the flexibility of some autolysis sites in bovine trypsin but not in salmon trypsin (experimental data) Design of “wet” experiments: site-directed mutagenesis Papaleo E., Riccardi L., Villa C., Fantucci P., De Gioia L., Biochim. Biophys. Acta, 1764 (2006) 1397. Acknowledgments Department of Biotechnology and Biosciences, University of Milano-Bicocca, Milano, Italy • Elena Papaleo • Prof. Piercarlo Fantucci • Chiara Villa, Laura Riccardi, Marco Pasi, Rodolfo Gonella Diaza, Paolo Mereghetti, Gianluca Santarossa Department of Biochemistry, University La Sapienza, Roma, Italy • Prof. Stefano Pascarella • Giulio Gianese • Daniele Tronelli • Prof. Arne Smalas Norwegian Structural Biology Centre, Tromso University, Tromso, Norway • Bjorn Bransdal • Magne Olufsen