* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Molecules, Genes, and Diseases Session 2 Protein Structure and
Signal transduction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Point mutation wikipedia , lookup
Expression vector wikipedia , lookup
Gene expression wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
Genetic code wikipedia , lookup
Interactome wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein purification wikipedia , lookup
Homology modeling wikipedia , lookup
Western blot wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
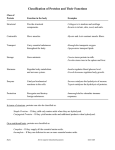
Molecules, Genes, and Diseases Sun 23/2/2014 Session 2 Protein Structure and Folding Dr. Mona A. Rasheed Structure of Session 2 Lecture 3: Protein Folding and Function 8:00 – 8:50 Lecture 4: Oxygen Transport Proteins (Hemoglobin & Myoglobin) 8:50 – 9:40 Work Session 1 10:00 – 12:00 MGD Session 2, Protein Structure & Function Learning Outcomes At the end of this session you should be able to: 1. Describe what is meant by the primary, secondary, tertiary and quaternary structure of proteins. 2. Describe the types of bonds and forces involved in protein structure. 3. Explain the key features of the two major secondary structure elements of proteins (α-helix and β -sheet). 4. Explain the physiological roles of myoglobin and haemoglobin. MGD Session 2, Protein Structure & Function Learning Outcomes 5. Contrast the oxygen-binding properties of myoglobin and haemoglobin and explain why haemoglobin is most suited to its role as an oxygen transporter. 6. Describe the major structural differences between oxygenated and deoxygenated haemoglobin and the molecular basis of cooperativity. 7. Describe the effects of CO2, H+, 2’3bisphosphoglycerate and carbon monoxide on the binding of oxygen by haemoglobin, and the physiological significance of these effects. 8. Appreciate that mutations in globin genes can give rise to diseases such as sickle cell anaemia or thalassemia. MGD Session 2, Protein Structure & Function Proteins • Proteins are the primary structural and functional polymers in living systems. • Only 20 amino acids are coded by DNA to appear in proteins. • Proteins are synthesized as a sequence of amino acids linked together in a linear polyamide (polypeptide) structure, but they assume complex three-dimensional shapes in performing their function. MGD Session 2, Protein Structure & Function Proteins • The linear sequence of the linked amino acids contains the information necessary to generate a protein molecule with a unique threedimensional shape. • The complexity of protein structure is best analyzed by considering the molecule in terms of four organizational levels, namely , primary, secondary , tertiary , and quaternary. MGD Session 2, Protein Structure & Function Proteins • Many proteins also contain modified amino acids and accessory components, termed prosthetic groups. • A range of chemical techniques is used to isolate and characterize proteins by a variety of criteria, including mass, charge, and threedimensional structure. MGD Session 2, Protein Structure & Function Levels of protein structure MGD Session 2, Protein Structure & Function PRIMARY STRUCTUR E OF PROTEINS • The sequence of amino acids in a protein is called the primary structure o f the protein . • Understanding the primary structure of proteins is important because many genetic disease s result in protein s with abnormal amino acid sequences , which cause improper folding and los s or impairment o f normal function. • The primary structures of the normal and the mutated protein s are known, this information ma y be used to diagnose or study the disease . MGD Session 2, Protein Structure & Function PRIMARY STRUCTUR E OF PROTEINS • The bonds responsible for the stabilization of primary structure is only the peptide bonds. • Peptide bond s are not broken by heating or high concentrations of urea. • Prolonged exposure t o a strong acid or base at elevated temperatures is required t o hydrolyze these bonds. MGD Session 2, Protein Structure & Function Secondary Structure of Proteins • The polypeptide backbone forms regular arrangements of amino acids that are located near to each other in the linear sequence. • These arrangements are termed the secondary structure of the polypeptide . • The α-helix, β-sheet , and β-bend are examples o f secondary structures. • Collagen helix , another example o f secondary structure , will discussed in the future. MGD Session 2, Protein Structure & Function Alpha Helix • The Alpha Helix Is a Coiled Structure Stabilized by Intra- chain Hydrogen Bonds. • Essentially all a helices found in proteins are right handed. • Hydrogen-Bonding Scheme For an a helix. In the a helix, the CO group of residue n forms a hydrogen bond with the NH group of residue n+ 4 MGD Session 2, Protein Structure & Function Alpha Helix MGD Session 2, Protein Structure & Function Alpha Helix MGD Session 2, Protein Structure & Function Alpha Helix Amino acids per turn Each turn o f a n α-helix contains 3.6 amino acids, and has a 0.54nm pitch. Thus , amino acid residues spaced three or four apart in the primary sequence are spatially close together when folded in the α-helix. Amin o acids that disrupt an α-helix : • Proline. • Large numbers of charged amino acid. • Large numbers of amino acids with bulky side chains, such as tryptophan , or amino acids, such as valine or isoleucine, that branch at the β-carbon . MGD Session 2, Protein Structure & Function β- Sheet 1. An Antiparallel β Sheet. Adjacent β strands run in opposite directions. Hydrogen bonds between NH and CO groups connect each amino acid to a single amino acid on an adjacent strand, stabilizing the structure. 16 Structure of a β - Strand. The side chains (green) are alternately above and below the plane of the strand 1. An Antiparallel β Sheet. 2. A Parallel β Sheet Adjacent β - strands run in the same direction. Hydrogen bonds connect each amino acid on one strand with two different amino acids on the adjacent strand. 3. Reverse Turn: The CO group of residue i of the polypeptide chain is hydrogen bonded to the NH group of residue i + 3 to stabilize the turn 20 Reverse Turn 21 Tertiary structure • The overall 3-dimensional structure of a protein is referred to as the tertiary structure. This involves folding up of the secondary structures so that amino acids far apart in the primary sequence may interact. • Larger proteins (~200 amino acids or greater) tend to have distinct domains. These are regions of the polypeptide that have distinct structures and often serve particular roles (e.g. ligand binding, interaction with other proteins etc.) 22 Tertiary structure • • • • • Bonds involved: Hydrogen bonds Van der Waals Hydrophobic interactions Covalent (disulphide) bonds Ionic interactions 23 24 25 Protein folding • Interactions between the side chains o f amino acids determine how a long polypeptide chain folds into the complex three-dimensional shape o f the functional protein . • Protein folding, occurs within the cell in seconds to minutes 26 Role o f chaperones in protein folding • The information needed for correct protein folding is contained in the primary structure of the polypeptide. • Why most proteins when denatured (see below) do not take up again their native conformations under favorable environmental conditions? 27 Role o f chaperones in protein folding • One answer to this problem is that a protein begins to fold in stages during it s synthesis , rather than waiting for synthesis o f the entire chain to be totally completed. • In addition, a specialized group o f proteins, named "chaperones," are required for the proper folding of many species of proteins. 28 Quaternary structure • Many proteins consist of more than 1 polypeptide chain. The polypeptide chains may be identical (homomeric proteins) or different (heteromeric proteins). • The arrangement of these subunits in such proteins is referred to as the quaternary structure. • The same types of bonds, that involved in tertiary structure, are involved in Quaternary structure. 29 Globular and fibrous proteins • Proteins can be categorised into 2 major groups depending of their higher order structure: globular or fibrous. • Most enzymes and regulatory proteins inside a cell tend to be globular proteins whereas fibrous proteins tend to provide structure, support and protection. 30 Protein Denaturation • The loss of protein structure sufficient to cause the loss of function is known as denaturation. • Denaturation is brought about by breaking the bonds that hold that maintain the protein’s tertiary and secondary structure. • Denaturing agents include heat, organic solvents, mechanical mixing, strong acids or bases , detergents, and ion s of heavy metals such as lead and mercury. 31 Role of b -Mercaptoethanol in Reducing Disulfide Bonds. Note that, as the disulfides are reduced, the b-mercaptoethanol is oxidized and forms dimers 32 Reduction and Denaturation of Ribonuclease 33 34