* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Support vector machines for protein function prediction
Immunoprecipitation wikipedia , lookup
Implicit solvation wikipedia , lookup
Structural alignment wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein design wikipedia , lookup
Circular dichroism wikipedia , lookup
List of types of proteins wikipedia , lookup
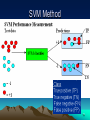
Protein folding wikipedia , lookup
Protein domain wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Homology modeling wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein purification wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Western blot wikipedia , lookup
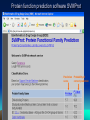
LSM3241: Bioinformatics and Biocomputing Lecture 3: Machine learning method for protein function prediction Prof. Chen Yu Zong Tel: 6516-6877 Email: [email protected] http://bidd.nus.edu.sg Room 07-24, level 7, SOC1, National University of Singapore Protein Function and Functional Family Proteins of similar functional characteristics can be grouped into a family 2 Protein Function and Functional Family Proteins of similar functional characteristics can be grouped into a family 3 Protein Function and Functional Family Proteins of similar functional characteristics can be grouped into a family 4 Functional Classification of Proteins by SVM • A protein is classified as either belong (+) or not belong (-) to a functional family • By screening against all families, the function of this protein can be identified (example: SVMProt) Protein Family-1 SVM - Family-2 SVM - Family-3 SVM + Protein belongs to Family-3 5 Functional Classification of Proteins by SVM What is SVM? • Support vector machines, a machine learning method, learning by examples, statistical learning, classify objects into one of the two classes. Advantages of SVM: • Diversity of class members (no racial discrimination). • Use of sequence-derived physico-chemical features as basis for classification. • Suitable for functional classification of novel proteins (distantly-related proteins, homologous proteins of different functions). 6 Machine Learning Method Inductive learning: Example-based learning Descriptor Positive examples Negative examples 7 Machine Learning Method Feature vectors: A=(1, 1, 1) B=(0, 1, 1) C=(1, 1, 1) D=(0, 1, 1) E=(0, 0, 0) F=(1, 0, 1) Descriptor Feature vector Positive examples Negative examples 8 SVM Method Feature vectors in input space: Z Input space Feature vector A=(1, 1, 1) B=(0, 1, 1) C=(1, 1, 1) D=(0, 1, 1) E=(0, 0, 0) F=(1, 0, 1) F E A B Y X 9 SVM Method Protein family members Border New border Protein family members Nonmembers Nonmembers Project to a higher dimensional space 10 SVM method New border Support vector Support vector Protein family members Nonmembers 11 SVM Method Support vector Protein family members Nonmembers New border Support vector 12 SVM Method Border line is nonlinear 13 SVM method Non-linear transformation: use of kernel function 14 SVM method Non-linear transformation 15 SVM Method 16 SVM Method 17 SVM Method 18 SVM Method 19 SVM for Classification of Proteins How to represent a protein? • Each sequence represented by specific feature vector assembled from encoded representations of tabulated residue properties: – amino acid composition – Hydrophobicity – normalized Van der Waals volume – polarity, – Polarizability – Charge – surface tension – secondary structure – solvent accessibility • Three descriptors, composition (C), transition (T), and distribution (D), are used to describe global composition of each of these properties. Nucleic Acids Res., 31: 3692-369720 SVM for Classification of Proteins How to represent a protein? 21 SVM for Classification of Proteins How to represent a protein? From protein sequence: To Feature vector : (C_amino acid composition, T_ amino acid composition, D_ amino acid composition, C_hydrophobicity, T_hydrophobicity, D_hydrophobicity, … ) Nucleic Acids Res., 31: 3692-3697 22 Protein function prediction software SVMProt Useful for functional prediction of novel proteins, distantly-related proteins, homologous proteins of different functions Your protein sequence Option 1 Your protein sequence Option 2 http://jing.cz3.nus.edu.sg/cgi-bin/svmprot.cgi Which functional families your protein belong to? Send sequence to classifier Input sequence through internet Computer loaded with SVMProt Input sequence on local machine Protein functional indications Support vector machines classifier for every protein functional family Identified Functional families Nucl. Acids Res. 31, 3692-3697 (2003) Protein function prediction software SVMProt Useful for functional prediction of novel proteins, distantly-related proteins, homologous proteins of different functions. Protein families covered: 46 enzyme families, 3 receptor families, 4 transporter and channel families, 6 DNA- and RNA-binding families, 8 structural families, 2 regulator/factor families. SVMProt web-version at: http://jing.cz3.nus.edu.sg/cgi-bin/svmprot.cgi Nucl. Acids Res. 31, 3692-3697 (2003) Protein function prediction software SVMProt Check covered protein families here Check format here Input sequence here Nucl. Acids Res. 31, 3692-3697 (2003) Protein function prediction software SVMProt Prediction Probability of score correct prediction Nucl. Acids Res. 31, 3692-3697 (2003) Summary of Today’s lecture • Machine learning method for protein function prediction. • Use of SVMProt for probing protein function 27