* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Introduction to Development
Genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression programming wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genomic imprinting wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genome evolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Designer baby wikipedia , lookup
Minimal genome wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetics of human development wikipedia , lookup
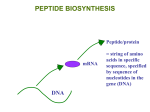
Molecular and Organismal Development Chapter 21: pp 411-429 Chapter 47: pp 992-1008 Topics in Development • 1. totipotency: development depends on selective expression of the whole genome present in every cell. • 2. blastula to gastrula: comparative analysis yields insights into the general nature of development • 3. the three fundamental processes: – cell division (differential rates of division are critical, programmed cell death is significant) – cell differentiation (changes in integration and shape are critical; targeting cells with signals is a critical part of the process) – morphogenesis of tissues and organs (includes defining the individual’s polarities, dividing the organism into segments, and – in animals -- migration of cells in tissue origin) Figure 21.2 Some key stages of development in animals and plants Figure 21.5 Test-tube cloning of carrots Figure 21.8 Working with stem cells Topics in Development • 1. totipotency: development depends on selective expression of the whole genome present in every cell. • 2. blastula to gastrula: comparative analysis yields insights into the general nature of development • 3. the three fundamental processes: – cell division (differential rates of division are critical, programmed cell death is significant) – cell differentiation (changes in integration and shape are critical; targeting cells with signals is a critical part of the process) – morphogenesis of tissues and organs (includes defining the individual’s polarities, dividing the organism into segments, and – in animals -- migration of cells in tissue origin) Figure 47.6 Cleavage in an echinoderm (sea urchin) embryo Figure 47.9 Sea urchin gastrulation (Layer 3) Figure 47.8x Cleavage in a frog embryo Figure 47.8d Cross section of a frog blastula Figure 47.10 Gastrulation in a frog embryo Figure 47.12 Gastrulation in a frog embryo Figure 47.12 Cleavage, gastrulation, and early organogenesis in a chick embryo The cells in the three germ layers have defined fates in the adult: Topics in Development • 1. totipotency: development depends on selective expression of the whole genome present in every cell. • 2. blastula to gastrula: comparative analysis yields insights into the general nature of development • 3. the three fundamental processes: – cell division (differential rates of division are critical, programmed cell death is significant) – cell differentiation (changes in integration and shape are critical; targeting cells with signals is a critical part of the process) – morphogenesis of tissues and organs (includes defining the individual’s polarities, dividing the organism into segments, and – in animals -- migration of cells in tissue origin) Figure 21.4 Cell lineage in C. elegans C. elegans cell targeting C. elegans cell targeting Figure 47.14 Organogenesis in a frog embryo Figure 47.16 Change in cellular shape during morphogenesis Apoptosis in development Topics in Development 4. Homeotic genes a. the determination of appendage identity on fruitfly segments b. the evolution of form in segmented animals 4. Morphogenesis in Plants 5. Organ identity genes in flower development Figure 21.11 Key developmental events in the life cycle of Drosophila Figure 21.12 The effect of the bicoid gene, a maternal effect (egg-polarity) gene Drosophila Figure 21.13 Segmentation genes in Drosophila Figure 21.13 Homeotic mutations and abnormal pattern formation in Drosophila Homeotic genes:the DNA sequence of the gene (blue) contains a 180 bp sequence—the homeobox—(red) that is highly conserved. The homeodomain - 60 amino acids of the homeotic gene product that remain very similar in all proteins made by homeotic genes. Figure 17.7 The initiation of transcription at a eukaryotic promoter control of transcription in C.elegans lin-3 | tctctccctattcaatgcacctgtgtattttatgctggttttttcttgtgaccctgaa aactgtacacacaggtgttcttaccaatgtctcaggcatttttggaaaagta atattaagaaaattatacatattttcttgaatacgaaaaatttaaATGTTC GGTAAATCGATTCCTGAACGACTTCTAGTCGCATTT HLH-2 binding site NHR binding site EXON is in uppercase letters. Figure 19.10 Three of the major types of DNA-binding domains in transcription factors The homeobox is relatively constant because it has a precise job. The homeobox is relatively constant because it has a precise job. Figure 21.14 Homologous genes that affect pattern formation in a fruit fly and a mouse Topics in Development 4. Homeotic genes a. the determination of appendage identity on fruitfly segments b. the evolution of form in segmented animals 4. Morphogenesis in Plants 5. Organ identity genes in flower development Serial Homology of the Lobster CRUSTACEANS COMPARED - EVOLUTIONARY HOMOLOGY CRUSTACEANS COMPARED: EVOLUTIONARY TRANSFORMATION ROCK CRAB LOBSTER Figure 33.28 Horseshoe crabs, Limulus polyphemus Figure 33.27 A trilobite fossil Figure 33.x1 Insecta: beetle Figure 32.8 Animal phylogeny based on sequencing of SSU-rRNA lophophore Figure 32.13x Burgess Shale fossils Figure 32.13 A sample of some of the animals that evolved during the Cambrian explosion Porifera and Cnidaria are prominent in the Burgess Shale Annelid worms in the Burgess shale Anomalocaris hunts so are arthropods!! Figure 26.8 The Cambrian radiation of animals Causes of the radiation: 1. Atmospheric oxygen reaches sufficient levels. 525 Burgess Shale P R D D PL PL PL PE 2. Predator-prey relationships originate. 3. Homeobox genes evolve. (Ediacaran) Evolutionary changes in the timing of homeobox genes yield morphological change. Artemia, the brine shrimp Another representation of the sequence of homeotic gene expression in an arthropod. Notice the continued prominence of Antp, Ubx, and Abd paralogs. Timing of expression of the homeobox genes Antp, Ubx, and AbdA. Fading lines indicate weaker expression later in development. Number of homeobox paralogs increases in arthropods. This illustration also shows change in timing of gene expression. Amino acid sequences compared for the homeotic gene Ubx insects arthropods Dm - Fruitfly Tc - Beetle Jc - Butterfly Ak - Onychophoran Decides six legs versus more Topics in Development 4. Homeotic genes a. the determination of appendage identity on fruitfly segments b. the evolution of form in segmented animals 4. Morphogenesis in Plants 5. Organ identity genes in flower development Figure 35.13 Morphology of a winter twig Padua Botanical Garden Johann Wolfgang von Goethe 1749 - 1832 Goethe’s inspiration - the European Fan Palm at the Padua Botanical Garden SERIAL HOMOLOGY - GOETHE’S PALM Figure 35.17 The terminal bud and primary growth of a shoot Figure 21.20a Organ identity genes and pattern formation in flower development: Normal flower development from Campbell, 6th edition A set of homeotic genes, in different combinations, yield the different floral organs. A B C E Figure 21.20b Organ identity genes and pattern formation in flower development: In situ hybridization from Campbell, 6th edition The evolutionary determination of sepal and petal. A B2 B3 C B1 Topics in Development 4. Homeotic genes a. the determination of appendage identity on fruitfly segments b. the evolution of form in segmented animals 4. Morphogenesis in Plants 5. Organ identity genes in flower development