* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 27
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Point mutation wikipedia , lookup
Catalytic triad wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Proteolysis wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Peptide synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Metalloprotein wikipedia , lookup
Genetic code wikipedia , lookup
Citric acid cycle wikipedia , lookup
Biochemistry wikipedia , lookup
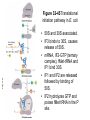
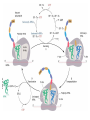
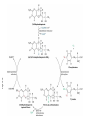
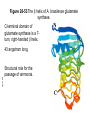
FCH 532 Lecture 25 Chapter 26: Amino acid metabolism Quiz Friday Glucogenic/Ketogenic amino acids (15 min) Quiz Monday April 2:Translation factors Exam 3 on Monday, April 9. Page 1323 Figure 32-45 Translational initiation pathway in E. coli. • 50S and 30S associated. • IF3 binds to 30S, causes release of 50S. • mRNA, IF2-GTP (ternary complex), fMet-tRNA and IF1 bind 30S. • IF1 and IF2 are released followed by binding of 50S. • IF2 hydrolyzes GTP and poises fMet tRNA in the P site. Page 1327 RF-1 = UAA RF-2 = UAA and UGA Cannot bind if EF-G is present. RF-3-GTP binds to RF1 after the release of the polypeptide. Hydrolysis of GTP on RF-3 facilitates the release of RF-1 (or RF-2). Page 1335 EF-G-GTP and ribosomal recycling factor (RRF)-bind to A site. Release of GDP-RF-3 EF-G hydrolyzes GTP -RRF moves to the P site to displace the tRNA. RRF and EF-G-GDP are released yielding inactive 70S Page 1322 Trp is both glucogenic and ketogenic • • • • Trp is broken down into Ala (pyruvate) and acetoacetate. First 4 reactions lead to Ala and 3hydroxyanthranilate. Reactions 5-9 convert 3-hydroxyanthranilate to aketoadipate. Reactions 10-16 are catalyzed by enzymes of reactions 5 - 11 in Lys degradation to yield acetoacetate. Page 1007 Page 1007 1. Tryptophan-2,3-dioxygenase, 2. Formamidase, 3. Kynurenine-3-monooxygense, 4. kynureninase (PLP dependent) • • • • Kynureinase, another PLP mechanism Reaction 4: cleavage of 3-hydroxykynurenine to alanine and 3-hydroxyanthranilate is catalyzed by the PLP dependent enzyme kynureinase. This facilitates a C-C bond cleavage. (previous reactions catalyzed the C-H and C-C bond cleavage) Follows the same steps as transamination but does not hydrolyze the tautomerized Schiff base. Enzyme amino acid acts as a nucleophile tto attack the carbonyl carbon (Cof the tautomerized 3hydroxykynurenine-PLP Schiff base. Page 1008 Page 1007 6. Amino carboxymuconate semialdehyde decarboxylase 7. Aminomuconate semialdehyde dehydrogenase 8. Hydratase, 9. Dehydrogense 10-16. Reactions 5-11 in lysine degradation. -keto acid dehydrogenase • Glutaryl-CoA dehydrogenase • Decarboxylase • Enoyl-CoA hydratase • -hydroxyacyl-CoA dehydrogenase • HMG-CoA synthase • Page 1006 • HMG-CoA lyase Phe and Tyr are degraded to fumarate and acetoacetate • • The first step in Phe degradation is conversion to Tyr so both amino acids are degraded by the same pathway. 6 reactions 1. 2. 3. Page 1009 4. 5. 6. Phenylanalnine hydroxylase Aminotransferase p-hydroxyphenylpyruvate dioxygenase Homogentisate dioxygenase Maleylacetoacetate isomerase Fumarylacetoacetase Phenylalanine hydroxylase has biopterin cofactor • • • • 1st reaction is a hydroxylation reaction by phenylalanine hydroxylase (PAH), a non-heme-iron containing homotetrameric enzyme. Requires O2, FeII, and biopterin a pterin derivative. Pterins have a pteridine ring (similar to flavins) Folate derivatives (THF) also contain pterin rings. Page 1009 Figure 26-27 The pteridine ring, the nucleus of biopterin and folate. Active BH4 must be regenerated • • • • • Active form in PAH is 5,6,7,8-tetrahydrobiopterin (BH4) Produced from 7,8-dihydrobiopterin via dihydrofolate reductase (NADPH dependent). 5,6,7,8-tetrahydrobiopterin is hydroxylated to pterin-4acabinolamine by phenylalanine hydroxylase. pterin-4a-cabinolamine is converted to 7,8dihydrobiopterin (quinoid form) by pterin-4a-carbinoline dehydratase 7,8-dihydrobiopterin (quinoid form) is reduced by dihydropteridine reductase to regenerate the active cofactor. Page 1010 NIH shift • • • A 3H that starts on C4 of Phe’s ring ends up on C3 of Tyr’s ring rather than being lost to solvent. Mechanism is called the NIH shift 1st characterized by scientists at NIH 1 and 2: activation of the enzyme’s BH4 and Fe(II) cofactors to yield pterin-4acarbinolamine and a reactive oxyferryl [Fe(IV)=O2-] 3: Fe(IV)=O2- reacts with Phe to form an epoxide across the 3,4 bond. 4: epoxide opening to form carbocation at C3 5: migration of hydride from C4 to C3 to form more stable carbocation. 6: ring aromatization to form Tyr Phe and Tyr are degraded to fumarate and acetoacetate • • • • The first step in Phe degradation is conversion to Tyr so both amino acids are degraded by the same pathway. 6 reactions Reaction 1 = 1st NIH shift Reaction 3 is also an example of NIH shift (26-31) 1. 2. 3. Page 1009 4. 5. 6. Phenylanalnine hydroxylase Aminotransferase p-hydroxyphenylpyruvate dioxygenase Homogentisate dioxygenase Maleylacetoacetate isomerase Fumarylacetoacetase Amino acid biosynthesis • • Essential amino acids - amino acids that can only be synthesized in plants and microorganisms. Nonessential amino acids - amino acids that can be synthesized in mammals from common intermediates. Page 1030 Table 26-2 Essential and Nonessential Amino Acids in Humans. Nonessential amino acid biosynthesis • • • Except for Tyr, pathways are simple Derived from pyruvate, oxaloacetate, -ketoglutarate, and 3phosphoglycerate. Tyrosine is misclassified as nonessential since it is derived from the essential amino acid, Phe. Glutamate biosynthesis • • • • • • • Glu synthesized by Glutamate synthase. Occurs only in microorganisms, plants, and lower animals. Converts -ketoglutarate and ammonia from glutamine to glutamate. Reductive amination requires electrons from either NADPH or ferredoxin (organism dependent). NADPH-dependent glutamine synthase from Azospirillum brasilense is the best characterized enzyme. Heterotetramer (22) with FAD, 2[4Fe-4S] clusters on the subunit and FMN and [3Fe-4S] cluster on the subunit NADPH + H+ + glutamine + -ketoglutarate 2 glutamate + NADP+ Figure 26-51 The sequence of reactions catalyzed by glutamate synthase. Electrons are transferred from NADPH to FAD at active site 1 on the subunit to yield FADH2. 2. Electrons transferred from FADH2 to FMN on site 2 to yield FMNH2. 3. Gln is hydrolyzed to glutamate and ammonia on site 3 of the subunit. 4. Ammonia is transferred to site 2 to form -iminoglutarate from -KG Page 1031 1. 5. -iminoglutarate is reduced by FMNH2 to form glutamate. Page 1032 Figure 26-52 X-Ray structure of the subunit of A. brasilense glutamate synthase as represented by its C backbone. Figure 26-53 The helix of A. brasilense glutamate synthase. C-terminal domain of glutamate synthase is a 7turn, right-handed helix. Page 1032 43 angstrom long. Structural role for the passage of ammonia. Ala, Asn, Asp, Glu, and Gln are synthesized from pyruvate, oxaloacetate, and -ketoglutarate • • • Pyruvate is the precursor to Ala Oxaloacetate is the precursor to Asp -ketoglutarate is the precursor to Glu Asn and Gln are synthesized from Asp and Glu by amidation. Page 1033 Figure 26-54 The syntheses of alanine, aspartate, glutamate, asparagine, and glutamine. Gln and Asn synthetases • • • • • Glutamine synthetase catalyzes the formation of glutamine in an ATP dependent manner (ATP to ADP + Pi). Makes glutamylphosphate intermediate. NH4+ is the amino group donor. Asparagine synthetase uses glutamine as the amino donor. Hydrolyzes ATP to AMP + PPi Glutamine synthetase is a central control point in nitrogen metabolism • • • • • Gln is an amino donor for many biosynthetic products and also a storage compound for excess ammonia. Mammalian glutamine synthetase is activated by ketoglutarate. Bacterial glutamine synthetase has more complicated regulation. 12 identical subunits, 469-aa, D6 symmetry. Regulated by different effectors and covalent modification. Figure 26-55a X-Ray structure of S. typhimurium glutamine synthetase. (a) View down the 6-fold axis showing only the six subunits of the upper ring. Active sites shown w/ Mn2+ ions (Mg2+) Adenylation site is indicated in yellow (Tyr) Page 1034 ADP is shown in cyan and phosphinothricin is shown (Glu inhibitor) Page 1034 Figure 26-55b Side view of glutamine synthetase along one of the enzyme’s 2-fold axes showing only the eight nearest subunits. Glutamine synthetase regulation • • • • • 9 feedback inhibitors control the activity of bacterial glutamine synthetase His, Trp, carbamoyl phosphate, glucosamine-6-phosphate, AMP and CTP-pathways leading away from Gln Ala, Ser, Gly-reflect cell’s N level Ala, Ser, Gly, are competitive with Glu for the binding site. AMP and CTP are competitive with the ATP binding site. Glutamine synthetase regulation • • • • • • E. coli glutmine synthetase is covalently modified by adenylation of a Tyr. Increases susceptiblity to feedback inhibition and decreases activity dependent on adenylation. Adenylation and deadenylation are catalyzed by adenylyltransferase in complex with a tetrameric regulatory protein, PII. Adensyltransferase deadenylates glutamine synthetase when PII is uridylated. Adenylates glutamine synthetase when PII lacks UM residues. PII uridylation depends on the activities of a uridylyltransferase and uridylyl-removing enzyme that hydrolyzes uridylyl groups. Glutamine synthetase regulation • • • Uridylyltransferase is activated by -ketoglutarate and ATP. Uridylyltransferase is inhibited by glutamine and Pi. Uridylyl-removing enzyme is insensitive to these compounds. Page 1035 Figure 26-56 The regulation of bacterial glutamine synthetase.