* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download File - Elko Science
Theories of general anaesthetic action wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Signal transduction wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein (nutrient) wikipedia , lookup
List of types of proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein domain wikipedia , lookup
Circular dichroism wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein folding wikipedia , lookup
Protein–protein interaction wikipedia , lookup
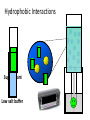
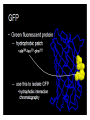
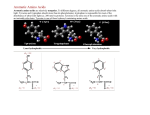
Intended Learning Objectives You should be able to… 1. Give 3 examples of proteins that are important to humans and are currently produced by transgenic organisms. 2. Describe a protein’s composition. 3. Recognize the 4 basic protein structures. 4. Explain the following key terms: Hydrophobic and Hydrophilic. 5. Honors: Identify the parts of an amino acid. Resources: Chapter 3 (3.14) and Notes Intended Learning Objectives You should be able to… 6. Explain how we will be able to isolate the GFP protein for purification. Resources: Lab Handouts and Notes Intended Learning Objectives You should be able to… 6a. Explain what each of the following do in order to help extract the GFP. - Lysozyme - Freezer - Centrifuge (1st and 2nd uses) Intended Learning Objectives You should be able to… 6b. Explain what each buffer did to the proteins in the column. Be sure to indicate the concentration of salt in each. - Equilibration buffer - Binding buffer - Wash buffer - Elution buffer Hydrophobic Interactions Supernatant Low salt buffer Protein Structure o 1 Primary o 2 Secondary o 3 Tertiary o 4 Quaternary 1° Primary Structure • Linear sequence of amino acids that make up the polypeptide chain. • Determined by the genetic code in particular the mRNA • The bond between two amino acids is a peptide bond. This bond is formed by the removal of a H2O molecule from two different amino acids, forming a dipeptide. • The sequence of amino acids determines the positioning of the different chemical groups of each building block which determines which groups will interact with each other and therefore determines the final structure and function of the molecule. 2° The secondary structure • coiling of the peptide chain into a helix or some other regular pattern of twists or kinks of the polypeptide chain. • due to hydrogen bonds forming between the atoms of the amino acid backbone of the polypeptide chain. • The two most common types of secondary structure are called the alpha helix and beta pleated sheet. 3° Tertiary structure • three dimensional globular structure formed by bending and twisting of the polypeptide chain. •folding of the polypeptide chain is stabilized by multiple weak, non-covalent interactions. These interactions include: + Hydrogen bonds - form when a Hydrogen atom is shared by two other atoms. + Electrostatic interactions - occur between charged amino acid side chains. + Hydrophobic interactions - amino acids with a polar (water soluble) side chain are often found on the surface of the molecule while amino acids with nonpolar (water insoluble) side chain are buried in the interior. This means that the folded protein is soluble in water or aqueous solutions. + Disulfide Bonds: The polypeptide chains of some proteins are linked by disulfide bonds. 4° Quaternary structure. • Proteins that contain more than one polypeptide chain • Each polypeptide chain in the protein is called a subunit. • The subunits can be the same polypeptide chain or different ones. • For some proteins, quaternary structure is required for full activity (function) of the protein. Hydrophobic interaction chromatography HIC The background – hydrophobic compounds stick together – association affected by: salt concentration greater concentration = more hydrophobic interactions Hydrophobic region The idea – stick GFP to a polymer sphere with hydrophobic groups – wash it off by changing the salt concentration ( decrease or increase?) True or False 1. The function of a protein is determined by its structure/shape. TRUE 2. Hydrophobic molecules tend to “hang out” or bind to other hydrophobic molecules. TRUE 3. Higher concentrations of salt decrease the binding of proteins to hydrophobic polymers.FALSE 4. Molecules that are Hydrophobic are polar molecules. FALSE 5. Electrostatic attractions between amino acids occur in the primary structure of a protein. FALSE Which of the following is the strongest bond? • Polar covalent • Nonpolar covalent • Hydrogen • Electrostatic Which protein structure does the picture depict below? a-helix or beta sheet? a-helix Primary, secondary, tertiary, or quaternary structure? secondary Which of the following weakened the cell wall of the bacteria cells. • lysozyme • freezer • centrifuge • arabinose Which of the following had the highest salt concentration? • Wash buffer • Equilibration buffer • Elution Buffer • Binding Buffer Hydrophobic Interaction Chromatography Biopolymer (phenyl agarose - Binding Surface) Driving force for hydrophobic adsorption Water molecules surround the analyte and the binding surface. When a hydrophobic region of a biopolymer binds to the surface of a mildly hydrophobic stationary phase, hydrophilic water molecules are effectively released from the surrounding hydrophobic areas causing a thermodynamically favorable change in entropy. Temperature plays a strong role Hydrophobic region Ammonium sulfate, by virtue of its good salting-out properties and high solubility in water is used as an eluting buffer (Christian G. Huber, Biopolymer Chromatography, Encylcopedia in analytical chemistry, 2000) Hydrophobic interaction chromatography (HIC) separates molecules based on their hydrophobicity. Molecules that contain both hydrophobic and hydrophilic regions are applied to an HIC column in a high-salt buffer. The salt in the buffer reduces the solvation of the sample. As overall solvation decreases, hydrophobic regions that become exposed are adsorbed by the medium. The more hydrophobic the molecule, the less salt is needed to promote binding. Usually a decreasing salt gradient is used to elute samples from the column in order of increasing hydrophobicity.