* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Introduction to Protein Science Architecture, Function
Lipid signaling wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Paracrine signalling wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Gene expression wikipedia , lookup
Genetic code wikipedia , lookup
Magnesium transporter wikipedia , lookup
Expression vector wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Biochemistry wikipedia , lookup
Interactome wikipedia , lookup
Western blot wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Homology modeling wikipedia , lookup
Protein purification wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
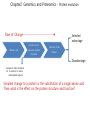
Introduction to Protein Science Architecture, Function, and Genomics Arthur M. Lesk Chapter 2: Genomics and Proteomics P47-51 2008-11-10 Jeong, Da-Geum, UST Chapter2: Genomics and Proteomics - Protein evolution Flow of Change----------------------------- 2.Amino acid or 1.Nucleic acid expression pattern of protein Selected advantage 3.Activity of the Protein Disadvantage Exception: Silent mutation Ex: 3rd position in exons, untranslated regions Simplest change to a protein is the substitution of a single amino acid Then, what is the effect on the protein structure and function? Chapter2: Genomics and Proteomics - Protein evolution 2. Change of the Structure(folding) 1.No noticeable effect Ex) Hemoglobin b-chain mutation -> Robust protein to mutation B60Val-> Glu, - rapid degraded Ex) enzymes in closely related species B106Leu->Arg - precipitate Substitution of (a single) amino acid 3.Sensitive position in the active site Ex) Gln-> Arg substitution, Malate dehydrogenase -> 4. Easy to aggregate-> Very serious clinical consequence Ex) Sickle cell anaemia, Z-mutant of a1-antitrypsin Lactate dehydrogenase Other type of change 1 Insertion 2 Deletion – cystic fibrosis 3 Transpositions – degraded in the ER Chapter2: Genomics and Proteomics - Protein evolution How do proteins develop new functions? (1) Divergence – progressive localized changes in sequence and structure -> initially to change in specificity -> ultimately to changes in the nature of the reaction catalysed (2) Recruitment – one protein is adapted (3)‘Mixing and matching’ of domains or modular evolution - large-scale structural changes - Individual domain-> gain of function, modified function, different processes. Robustness of protein structure to mutations is a maintenance of structure in spite of the divergence of sequences during evolution