* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Oxidative Phosphorylation - Study in Universal Science College
Proteolysis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Photosynthesis wikipedia , lookup
Mitochondrion wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Biochemistry wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Citric acid cycle wikipedia , lookup
Microbial metabolism wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Metalloprotein wikipedia , lookup
Electron transport chain wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
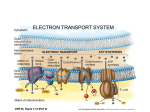
Oxidative Phosphorylation •It is the process by which electrons are carried from reduced cofactors (NADH+/ QH2) are finalled in stepwise manner to oxygen. •Electrons flow much like electricity in a circuit with free energy being conserved with the formation of proton gradient. • In the end the investment of reduced cofactors are utilized in the production of ATP •Reduced cofactors like NADH+ are produced during glycolysis, Citric acid cycle and fatty acid oxidation •During cellular respiration oxidative phosphorylation chemical energy of these reduced molecules are utilized to produce ATP •The ultimate acceptor of e- through a series of O/R reactions is the O2 within the mitochondrion Mitochondrial Anatomy • The anatomy of the mitochondrion reflects its role in oxidative phosphorylation • The outer membrane is porous and allows the free diffusion of molecules due to the presence of channel protein called porins • The inner membrane is impermeable to most substances including ions and encloses a space called the matrix • The inner membrane is convoluted structure which provides greater surface area for the protein complexes of the oxidative phosphorylation • The mitochondrion consists two membranes separated by a compartment called the intermembrane space • During oxidative phosphorylation protons are pumped into this compartment matrix cristae intermembrane space inner membrane mitochondrion outer membrane Transport Shuttles • Since the inner mitochondrial membrane is impermeable to most molecules, NADH+ produced by glycolysis in the cytosol must be imported into via biochemical reaction of the malate – aspartate shuttle • This shuttle operates mainly in the liver, kidney and heart • In the skeletal muscle & brain NADH+ is imported into - by Glycerol – phosphate shuttle • The process is a formal currency exchange between one region of the cell with the other. • ATP, ADP and Pi also require transport protein for their import and export across the inner membrane Redox potential of the components of Respiratory chain • In the respiratory chain the e- s are transferred from reducing equivalents to a chain of e- carriers which are arranged sequentially • The e- s flow from more electropositive components to more electronegative components i.e. to more positive re-dox potentials • Hydrogen and e- s move from NAD+/NADH to O2/ H2O Components of the Respiratory chain • The inner membrane consists of majority of proteins including three major electron transporting complexes • These complexes function in re oxidizing the coenzymes ( NAD+/ ubiquinone, etransferring flavoproteins) that have been reduced by dehydrogenases in the metabolic reactions within the mitochondrion • The terminal e- acceptor is O2 & the reaction is coupled to ATP synthesis Components of Respiratory chain • The respiratory chain consists of number of redox carriers proceeding from NAD – linked dehydrogenase through flavoprotein and cytochromes to molecular oxygen • Certain substrates (fumarate/succinate) comparatively of their more positive redox potential however are directly linked to flavoprotein dehydrogenase further linked to cytochromes to molecular oxygen • Ubiquinone (Q/ Coenzyme Q) links flavoprotein to Cytochrome b (member of cytochrome chain with lowest redox potential) • Ubiquinone acts as a mobile component of the respiratory chain that collects reducing equivalents from more fixed flavoproteins and passes them to cytochromes • Next component is the Iron – sulfur protein (Fe-S – a non heme protein) associated with flavoprotein and cytochrome b • Electrons flow through a series of cytochromes in order of increasing redox potentials to molecular oxygen • The terminal cytochrome aa3 (cytochrome oxidase) is responsible for the final combination of reducing equivalents with molecular oxygen. • It has a high affinity for O2 thus allowing the respiratory chain to function at its maximum. • This is the only irreversible reaction in the chain and hence provides direction to the movement of reducing equivalents & to the production of ATP – to which it is coupled • The components of the respiratory chain are all present in the inner mitochondrial membrane as four protein – lipid respiratory chain complexes • Cytochrome c is the only soluble cytochrome & together with ubiquinone seems to be a mobile component connecting the more fixed complexes In simple outline, ETC involves the removal of hydrogen atoms from the oxidizable substrates; these hydrogen atoms enter the ETC, a system of membrane – bound complexes and each soon split to yield a proton and electron. These electrons then pass through a series of cytochromes, finally reacting with molecular oxygen and the protons that were released earlier, to form water Electron carriers are Multi enzyme complexes • • • Complex I (NADH to Ubiquinone) Also k/as NADH:ubiquinone reductase Electron microscope reveals it as an L – shaped structure with one arm in the membrane and the other extending into the matrix • An enzyme with 42 polypeptide chains and an FMN – flavoprotein with about 6 Fe- S centres • Complex I catalyses 2 simultaneous reactions 1. - exergonic transfer of a hydride ion (:H-)from NADH & a proton from the matrix NADH + H+ + Q NAD+ + QH2 2. -endergonic transfer of 4 protons from the matrix to the intermembrane space Complex I – k/as proton pump driven by the energy of electron transfer; where – protons move from one location (matrix which then becomes negatively charged) to the other (intermembrane space which becomes positively charged) Structure of iron – sulfur center • These function as prosthetic groups which facilitate electron transfer • Iron - sulfur protein - The iron is not present in heme but is found in association with inorganic sulfur or the cysteine residues in the protein • Rieske Iron - sulfur protein - One iron atom is coordinated to 2 Histidine residues instead of 2 cysteine residues • All these centers however participate in one – electron transfer where the Fe –atom gets oxidised/ reduced • At least 8 Fe – S proteins are involved in mitochondrial electron trasfer Ubiquinone • Lipid soluble bezoquinone with a long isoprenoid side chain • Complete reduction of ubiquinone requires 2 electrons and 2 protons ( a 2 step rxn) through semiquinone as an intermediate • As it carries both e- & protons it plays a central role in coupling electron flow to proton movement • It always acts at the junction between a 2 e- donor and one electron acceptor O O CH3O CH 3 CH3O CH 3 CH3O (CH 2 CH O C CH 3 e CH 2)nH CH 3 CH3O (CH 2 CH O coenzyme Q C CH 2)nH coenzyme Q • e + 2 H+ OH CH3O CH 3 CH 3 CH3O (CH 2 CH OH C CH 2)nH coenzyme QH2 Complex II • Also called as Succinate:ubiquinone oxidoreductase (Succinate to ubiquinone) • It is the only membrane bound enzyme (succinate dehydrogenase) encountered in citric acid cycle • Structurally it is simpler than complex I with 2 types of prosthetic groups & 4 different proteins • One of the protein is bound covalently to FAD & Fe- S center with 4 Fe atoms • Electrons pass from succinate to FAD; through Fe – S centers finally reaching ubiquinone Oxidation of NADH & Succinate • NADH produced by CAC diffuses into the ETC where the flavoprotein enzyme (NADH dehydrogenase) oxidizes it to NAD+ • The process involves transfer of a Hydride ion (which consists of hydrogen nucleus with 2 associated electrons) to the flavin enzyme (FMN) which accepts the proton to be FMNH2 • To be known – Hydride ions do not have independent existence – they just represent the moiety transferred in biological reduction process • Succinate dehydrogenase – flavoprotein having FAD as the coenzyme – links CAC directly to ETC Fate of FMNH2 & FADH2 • FMNH2 & FADH2 are oxidized by enzymes that transfer the hydrogen atoms to a molecule of Ubiquinone (Q) thus forming Ubiquinol (QH2) • Ubiquinone also accepts hydrogen atoms transferred from other molecules that have been oxidized by ETC ( - oxidation, glycerol – 3 – P) Succinate dehydrogenase (fp) Fatty acyl CoA dehydrogenase (fp) Glycerol 3 phosphate dehydrogenase (fp) Matrix NADH fp Dehydrogenase Fes Q Q bc1 FeS Intermembrane space Proton pump Proton pump