* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Week 4
Metalloprotein wikipedia , lookup
Biochemistry wikipedia , lookup
Mitochondrion wikipedia , lookup
Photosynthesis wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Citric acid cycle wikipedia , lookup
Microbial metabolism wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Light-dependent reactions wikipedia , lookup
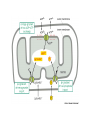
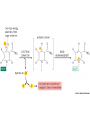
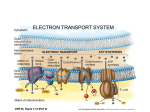
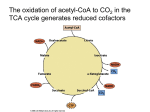
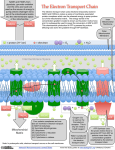
Week 4 Mitochondrion • See Figure 13-7 • Double membrane organelle • Outer membrane – large pores (channel-forming proteins permeable to molecules <MW=5000) – lipid metabolism • Inner membrane – convoluted to increase surface area – functions: electron transport chain; maintain proton gradient; transport proteins Mitochondrion • Intermembrane space – enzymes to phosphorylate nucleotides using ATP • Matrix – contains Kreb’s cycle (TCA or citric acid cycle) enzymes; fatty acid oxidation enzymes Endosymbiont theory • Theorizes about the origin of mitochondria and chloroplasts • Mitochondrion origins - aerobic, heterotrophic bacteria • Evidence: – – – – RNA Ribosomes antibiotic sensitivity DNA ATP formation • Driving force- electrochemical proton gradient = “proton-motive force” • Electrochemical gradient is made up of membrane potential + proton gradient (syn. pH gradient) • see Figure 13-12 Membrane potential • See Figure 13-2 • = matrix side negative and intermembrane space positive due to protons Proton gradient • See Figure 13-12 • = difference in proton concentration across inner membrane – pH of intermembrane space = 7 – pH of matrix = 8 • Remember - membranes are impermeable to protons ATP synthase • See Figures 13-14 & 15 • Two functions – ATP synthesis - synthesizes ATP using oxidative phosphorylation – ATP hydrolysis - pumps protons from matrix into intermembrane space to maintain electrochemical proton gradient Chemiosmotic coupling theory • Peter Mitchell, 1968 • theory: couple electron transport and proton pumping with ATP synthesis • electron transport generates membrane potential plus proton gradient (=proton motive force) • proton motive force drive ATP synthesis Other Roles • See Figure 13-16 • Proton gradient – pyruvate and phosphate import into matrix • Membrane potential – ADP-ATP exchange High energy electrons • Source – oxidation of organic compounds all heterotrophic organisms – light reactions in photosynthesis in photosynthetic eukaryotes and prokaryotes – oxidation of reduced inorganic compounds (e.g., NH4+ or H2S) in some prokaryotes • Carrier: NAD+ & FAD (see Figure 13-8) – NAD+ + 2e- + H+ ---> NADH Electron transport chain • See Figure 13-10 • Syn. Respiratory chain • embedded in inner membrane of mitochondrion • composed of 3 large complexes – NADH dehydrogenase – cytochrome b-c1 – cytochrome oxidase Electron transport chain • Begins with reduced NADH donating electron pair to NADH dehydrogenase complex • Electrons move from NADH dehydrogenase to cytochrome b-c1 to cytochrome oxidase to finally O2 to form water. – Why do electrons move in this fashion? – What drives protons out of matrix into intermembrane space? Why do electrons move in this fashion? • Depends on the relative affinity of the complexes for electrons. • Relative affinity is measured by the redox potential (E’o; mVolts) of the complex – negative potential = good electron donor – positive potential = good electron acceptor – this is all relative, e.g., -320 mV (good donor) vs -250 mV (good acceptor) • See panel 13-1 Redox potentials • NADH; - 320 mVolts • O2; +820 mVolts • electrons move from molecules with more negative redox potentials to more positive redox potentials What drives protons out of matrix into intermembrane space? • Energy released as electron moves down the electron transport chain. • Quantify the energy DGo = - n (0.023) (DE’o); n = number of electrons used; DE’o = difference in redox potential between oxidizing agent and reducing agent (see below) Calculating DGo • • • • NADH + O2 <----> NAD+ + H2O NAD+ + 2 e- + H+ ---> NADH (-320mV) O2 + 2e- + 2H+ ---> H2O (+820mV) Combine above half reactions: – NADH + O2 + 2e- + 2H+ ---> NAD+ + 2 e- + H+ + H2O – Cancel to get : NADH + O2 + H+ --> NAD+ + H2O DGo = - n (0.023) (DE’o) Calculating DGo DGo = - n (0.023) (DE’o) DE’o = E’o oxidizing agent - E’o reducing agent DE’o = 820 - (-) 320 mV = 1140 mV DGo = - 2 (0.023) (1140mV) DGo = - 52.4 kcal / mole •