* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Conformational Analysis of a Set of Peptides Corresponding
List of types of proteins wikipedia , lookup
Protein phosphorylation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein (nutrient) wikipedia , lookup
P-type ATPase wikipedia , lookup
Protein folding wikipedia , lookup
Homology modeling wikipedia , lookup
Protein domain wikipedia , lookup
Circular dichroism wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Protein structure prediction wikipedia , lookup
Proteolysis wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
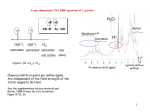
Conformational Analysis of a Set of Peptides Corresponding to the Entire Primary Sequence of the N-terminal Domain of the Ribosomal Protein L9: Evidence for Stable Native-like Secondary Structure in the Unfolded State Donna L. Luisi, Wen-Jin Wu and Daniel P. Raleigh* J. Mol. Biol. (1999) 287, 395-407. Speaker:Wu Chih-Wei Date : 2000/3/24 Introduction to Ribosome Ribosome contains one mRNA and two tRNA binding sites It contains two subunits 50S and 30S Structure of Ribosome Molecular biology of the cell. P.232 Schematic localization of L9 protein on the 50S ribosomal subunit J.B.C. (1991) 266. .33. 22129-22135. The structure of L9 protein Why Protein L9? L9 forms an interesting bilobal structure with a compact N-terminal domain connected by a long solvent-exposed α-helix to a compact C-terminal domain. Protein L9 does not appear to participate in subunit interaction nor in peptidyltransferase activity. L9 is one of the simplest examples of sheet-helix structures. Lack disulfide bond and no cofactors N-terminal domain of L9 CD spectra of the five peptides PH 5.4 , 4℃ 206nm 222nm β1:1-11 β2:12-23 α1: 24-34 β3:35-42 α2:40-56 CD standard curve α: αhelix β : β sheet Rc : random coil Far-UV CD spectra of α-1 □ : 75μM ● : 500 μM Below 100μM random coil 100-545 μM not random coil, (self-assocites) NMR spectrum of β-2 DQF-COSY spectrum ROESY spectrum The β-1, β-2, β-3 peptide are unstructure in solution. Summary of NMR data CSI : +1 (βsheet) 0 (no structure) –1 (α-helix) 3J HNCα:● : below 6 HZ (α-helix) ○ : above 8 HZ(β-sheet) - : not measure : 6-8 HZ(random coli) The α-2 Peptide NMR Spectrum PH 5.4 , 4℃ DQF-COSY spectrum NOESY spectrum Get 9 amino acid and 2 of 5 are too close to the diagonal. Provide more direct evidence for the helix formation. A native N-capping interaction stabilizes the helical structure α-2 : 40-56 (53% helix) α-2B: 41-56 (32% helix ) PH 5.4 4℃ CD spectra of the peptide α-2 and α-2B Conclusion The pH and ionic strength dependence of the helical content of α-2. The change in θ222 from 0.4M to 1.6M NaCl is less than 3%. The change in θ222 in PH 11 is less than 10%. The change in θ222 in PH 2 is more than 10%, corresponding to an apparent increase in helicity. Thr40 acts as an N-capping residue and its side-chain forms a H-bond with the amide proton of residue 43. The peptides β-3 and α-2 provide a model of cis-trans proline isomerism in the unfolded state. Trans CαThr40– Cδ Pro41 (native state) Cis CαThr40– Cα Pro41 Cis-trans proline isomerism ~The End