* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Susan - Stanford University
Immunoprecipitation wikipedia , lookup
Cooperative binding wikipedia , lookup
List of types of proteins wikipedia , lookup
Circular dichroism wikipedia , lookup
Implicit solvation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein design wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein domain wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Protein folding wikipedia , lookup
Structural alignment wikipedia , lookup
Homology modeling wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
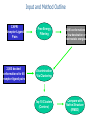
ClusPro: an automated docking and discrimination method for the prediction of protein complexes Stephen R. Comeau, David W.Gatchell, Sandor Vajda, and Carlos J. Camacho Bioinformatics Graduate Program and Department of Biomedical Engineering, Boston University Presentation by: Susan Tang Background & Motivation • • • • • • Docking = process of starting with a set of coordinates for two distinct molecules and generating a model of the bound complex Numerous methods which perform protein- protein docking exist today Fourier correlation approach (Ritchie and Kemp, 2000) enabled the generation of billions of possible docked conformation via defined scoring functions Problem: Many false-positives (good surface complementarity) that are far from the native complex Motivation: Need to develop methods to filter and rank the docked conformations such that near-native complexes can be identified ClusPro: an automated, fast rigid-body docking and discrimination algorithm that: 1) Rapidly filters docked conformations 2) Ranks the conformations using clustering of computed pairwise RMSD values Input and Method Outline CAPRI Receptor-Ligand Pairs 2,000 docked conformations for 48 receptor-ligand pairs Free Energy Filtering 2,000 conformations w/ low desolvation or electrostatic energies Discrimination Via Clustering Top 10 Clusters (Centers) Compare with Native Structure (RMSD) Part I: Free-Energy Filtering • • • • • • • Goal: to identify docked conformations having good surface complementarity by selecting those w/ lowest desolvation and electrostatic energies Surface complementarity is an important criteria due to the observation that proteins tend to bury large surface areas after complex formation Electrostatic and desolvation potentials (capturing the free energy of association) are used independently since different binding mechanisms are governed by different ratios of electrostatic/desolvation contributions 500 structures w/ lowest values of desolvation free energy retained 1500 structures w/lowest electrostatic energy retained Electrostatics more sensitive to small coordinate perturbations noisy Cannot combine desolvation and electrostatics due to the noisy behavior of electrostatics potential Part II: Clustering based on Pairwise RMSD • • • By examining free energy landscapes of partially solvated receptor-ligand complexes: native binding site is expected to be characterized by a local minima having greatest width In other words, the most probable conformation is expected to be surrounded by lots of other low-energy conformations Goal: to use a hierarchical clustering method to select and rank docked conformations having the most “neighbors” given a defined cluster radius (in terms of C-alpha RMSD) Procedure: 1) Need to define fixed molecule (receptor) and flexible molecule (ligand) 2) Define a set of relevant ligand residues to be within 10 Angs of any atom in receptor 3) For each docked conformation X, calculate its pairwise ligand RMSD with 1999 other conformations Pairwise ligand RMSD = deviations between coordinates of X’s defined set of ligand residues and corresponding coordinates of another conformation 4) Cluster the set of 2000 docked conformations using a 2000 by 2000 matrix of RMSD values, and a cluster radius constraint of 9 Angs RMSD from the center 5) Pick largest cluster rank cluster center remove conformations within this cluster from matrix 6) Pick next largest cluster -> rank cluster center remove conformations within this cluster from matrix keep iterating until matrix is empty Results Result I: • Tested the discrimination step of the method on a benchmark set of 48 interacting protein pairs (2000 docked conformations each) • In 31/48 protein pairs, top 10 predictions include at least one near-native complex (average RMSD of 5 angs from native structure) Result II: - Tested method in the CAPRI (Critical Assessment of Predictions of Interactions) experiment and generated predictions for 9 target complexes - Round 3 (automated server): ClusPro prediction ranked as #3 for Target 8 ClusPro Web Server • • • • • User Input: PDB files of the 2 protein structures that user would like to analyze in terms complex formation Output: 10 (default) top predictions of docked conformations closest to native structure First, docking of the 2 proteins is performed using 2 established FFT-based docking programs (DOT and ZDOCK) Then, filtering and discrimination is performed Server allows for customization of parameters: – Clustering radius Smaller protein smaller radius maybe more suitable – Relative number of desolvation and electrostatic best hits used during filtering – Number of predictions to generate (1-30) Protein Drug Discovery • • • • • • • • Although small molecule drugs are more prevalent therapeutics in current drug discovery, protein drugs is a rapidly growing area in pharmaceuticals It is true that protein therapeutics can be much more costly (in terms of R&D and synthesis) than small-molecule therapeutics, but protein therapeutics can deliver biological mechanisms that are not possible with small-molecule therapeutics Multiple blockbuster protein drugs are currently on the market Conservative estimation: there exist between 3,000 and 10,000 possible drug targets Many of these new targets offer great opportunities for the development of protein drugs In 2002, drug companies sold nearly $33 billion in protein drugs Rising at an average annual growth rate (AAGR) of 12.2%, this market is expected to reach $71 billion in 2008. Examples of popular classes of drug targets: 1) G-protein-coupled receptors Compounds will be screened for their ability to inhibit (antagonist) or stimulate (agonist) the receptor 2) Protein kinases Compounds will be screened for their ability to inhibit the kinase Application to Protein Drug Discovery • • Ideal Drug: demonstrate high specificity and high affinity for the target protein In order to evaluate the affinity of the potential drug with the target, you must first predict what the binding interface looks like, and the relative positions of the potential drug and target • ClusPro is the first integrated automated server that incorporates both docking and discrimination steps for structural predictions of protein-protein complexes Using ClusPro, one can generate many relative orientation/conformations of the 2 proteins filter using desolvation + electrostatics potentials discriminate via clustering find the best fit (closest to native structure from x-ray crystallography results) between the 2 proteins Top ranked predictions of ClusPro further manual refinement and discrimination using existing biochemical constraints and analysis to eliminate false positives test binding affinity of promising protein pairs in vitro lead compounds used as starting points for drug development/optimization • • • • Can use ClusPro to screen databases of various existing, recombinant, or de novo proteins for their interaction to a protein target of interest ClusPro can be used to predict either: – How a protein drug may bind (either inhibit or stimulate) a receptor – How 2 proteins bind, and based on the structural details of the interaction design/screen for a drug that can inhibit that interaction